Mol:FL5FAHGA0001
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 35 38 0 0 0 0 0 0 0 0999 V2000 | + | 35 38 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.0380 -5.1440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0380 -5.1440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5947 -5.0325 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5947 -5.0325 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8144 -4.4288 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8144 -4.4288 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4015 -3.9368 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4015 -3.9368 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2312 -4.0483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2312 -4.0483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4509 -4.6519 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4509 -4.6519 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6212 -3.3331 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6212 -3.3331 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2083 -2.8411 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2083 -2.8411 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4244 -2.9526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4244 -2.9526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6441 -3.5563 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6441 -3.5563 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1143 -3.2461 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1143 -3.2461 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8371 -2.4607 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8371 -2.4607 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6132 -1.8456 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6132 -1.8456 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0340 -1.3440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0340 -1.3440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6787 -1.4577 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6787 -1.4577 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9027 -2.0730 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9027 -2.0730 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4818 -2.5744 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4818 -2.5744 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2575 -5.7474 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2575 -5.7474 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6377 -2.1073 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6377 -2.1073 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0883 -0.7568 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 1.0883 -0.7568 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 0.7007 -1.4282 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 0.7007 -1.4282 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.4461 -1.2151 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4461 -1.2151 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1961 -1.4282 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.1961 -1.4282 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.5838 -0.7568 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.5838 -0.7568 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.8383 -0.9698 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 1.8383 -0.9698 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 0.3612 -0.9516 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3612 -0.9516 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1071 -0.5041 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1071 -0.5041 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5838 -0.1580 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5838 -0.1580 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3215 -0.6917 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3215 -0.6917 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4466 -4.3173 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4466 -4.3173 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8102 -0.7290 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8102 -0.7290 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6721 -1.4112 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6721 -1.4112 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3865 -1.8237 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3865 -1.8237 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5192 -1.8939 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5192 -1.8939 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4796 -1.6150 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4796 -1.6150 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 20 21 1 0 0 0 0 | + | 20 21 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 20 1 0 0 0 0 | + | 25 20 1 0 0 0 0 |
| − | 20 26 1 0 0 0 0 | + | 20 26 1 0 0 0 0 |
| − | 25 27 1 0 0 0 0 | + | 25 27 1 0 0 0 0 |
| − | 24 28 1 0 0 0 0 | + | 24 28 1 0 0 0 0 |
| − | 21 19 1 0 0 0 0 | + | 21 19 1 0 0 0 0 |
| − | 19 8 1 0 0 0 0 | + | 19 8 1 0 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 15 29 1 0 0 0 0 | + | 15 29 1 0 0 0 0 |
| − | 3 30 1 0 0 0 0 | + | 3 30 1 0 0 0 0 |
| − | 14 31 1 0 0 0 0 | + | 14 31 1 0 0 0 0 |
| − | 23 32 1 0 0 0 0 | + | 23 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 16 34 1 0 0 0 0 | + | 16 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 32 33 | + | M SAL 2 2 32 33 |
| − | M SBL 2 1 35 | + | M SBL 2 1 35 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SVB 2 35 2.6721 -1.4112 | + | M SVB 2 35 2.6721 -1.4112 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 34 35 | + | M SAL 1 2 34 35 |
| − | M SBL 1 1 37 | + | M SBL 1 1 37 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 37 0.8012 1.8237 | + | M SVB 1 37 0.8012 1.8237 |
| − | S SKP 8 | + | S SKP 8 |
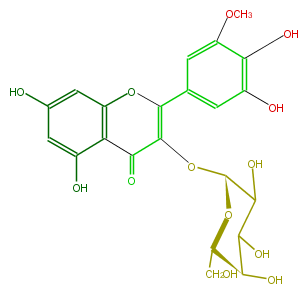
| − | ID FL5FAHGA0001 | + | ID FL5FAHGA0001 |
| − | KNApSAcK_ID C00005760 | + | KNApSAcK_ID C00005760 |
| − | NAME Laricitrin 3-galactoside | + | NAME Laricitrin 3-galactoside |
| − | CAS_RN 71393-05-0 | + | CAS_RN 71393-05-0 |
| − | FORMULA C22H22O13 | + | FORMULA C22H22O13 |
| − | EXACTMASS 494.10604078999995 | + | EXACTMASS 494.10604078999995 |
| − | AVERAGEMASS 494.40228 | + | AVERAGEMASS 494.40228 |
| − | SMILES c(C(=O)1)(c4O)c(cc(c4)O)OC(c(c3)cc(OC)c(O)c(O)3)=C1O[C@H](O2)C(O)C([C@H]([C@@H](CO)2)O)O | + | SMILES c(C(=O)1)(c4O)c(cc(c4)O)OC(c(c3)cc(OC)c(O)c(O)3)=C1O[C@H](O2)C(O)C([C@H]([C@@H](CO)2)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
35 38 0 0 0 0 0 0 0 0999 V2000
-0.0380 -5.1440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5947 -5.0325 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8144 -4.4288 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4015 -3.9368 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2312 -4.0483 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4509 -4.6519 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6212 -3.3331 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2083 -2.8411 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4244 -2.9526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6441 -3.5563 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1143 -3.2461 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8371 -2.4607 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6132 -1.8456 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0340 -1.3440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6787 -1.4577 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9027 -2.0730 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4818 -2.5744 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2575 -5.7474 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6377 -2.1073 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0883 -0.7568 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
0.7007 -1.4282 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.4461 -1.2151 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1961 -1.4282 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.5838 -0.7568 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.8383 -0.9698 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
0.3612 -0.9516 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1071 -0.5041 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5838 -0.1580 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3215 -0.6917 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4466 -4.3173 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8102 -0.7290 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6721 -1.4112 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3865 -1.8237 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5192 -1.8939 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4796 -1.6150 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
20 21 1 0 0 0 0
21 22 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 20 1 0 0 0 0
20 26 1 0 0 0 0
25 27 1 0 0 0 0
24 28 1 0 0 0 0
21 19 1 0 0 0 0
19 8 1 0 0 0 0
23 22 1 1 0 0 0
15 29 1 0 0 0 0
3 30 1 0 0 0 0
14 31 1 0 0 0 0
23 32 1 0 0 0 0
32 33 1 0 0 0 0
16 34 1 0 0 0 0
34 35 1 0 0 0 0
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 32 33
M SBL 2 1 35
M SMT 2 CH2OH
M SVB 2 35 2.6721 -1.4112
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 34 35
M SBL 1 1 37
M SMT 1 OCH3
M SVB 1 37 0.8012 1.8237
S SKP 8
ID FL5FAHGA0001
KNApSAcK_ID C00005760
NAME Laricitrin 3-galactoside
CAS_RN 71393-05-0
FORMULA C22H22O13
EXACTMASS 494.10604078999995
AVERAGEMASS 494.40228
SMILES c(C(=O)1)(c4O)c(cc(c4)O)OC(c(c3)cc(OC)c(O)c(O)3)=C1O[C@H](O2)C(O)C([C@H]([C@@H](CO)2)O)O
M END