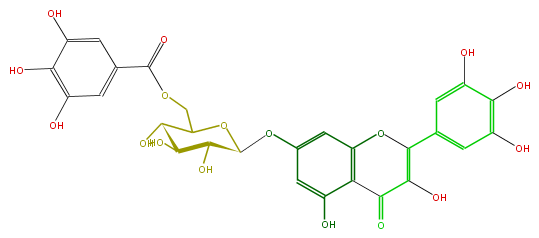
Mol:FL5FAGGS0032
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 45 49 0 0 0 0 0 0 0 0999 V2000 | + | 45 49 0 0 0 0 0 0 0 0999 V2000 |
| − | 4.9789 -0.7657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9789 -0.7657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2644 -0.3532 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2644 -0.3532 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2644 0.4718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2644 0.4718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9789 0.8843 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9789 0.8843 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.6934 0.4718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.6934 0.4718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.6934 -0.3532 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.6934 -0.3532 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5500 -0.7657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5500 -0.7657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8355 -0.3532 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8355 -0.3532 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1210 -0.7657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1210 -0.7657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1210 -1.5907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1210 -1.5907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8355 -2.0032 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8355 -2.0032 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5500 -1.5907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5500 -1.5907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4066 -0.3532 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4066 -0.3532 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6921 -0.7657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6921 -0.7657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6921 -1.5907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6921 -1.5907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4066 -2.0032 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4066 -2.0032 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8355 -2.7101 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8355 -2.7101 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0224 -0.3532 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0224 -0.3532 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2644 -2.0032 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2644 -2.0032 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4066 -2.6942 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4066 -2.6942 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.4079 0.8843 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.4079 0.8843 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9789 1.7093 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9789 1.7093 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.3713 -0.7446 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.3713 -0.7446 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7730 -0.1250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7730 -0.1250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3603 -0.8397 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3603 -0.8397 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5620 -0.6305 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5620 -0.6305 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7685 -0.8574 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7685 -0.8574 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1811 -0.1426 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1811 -0.1426 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9795 -0.3517 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9795 -0.3517 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1365 0.2341 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1365 0.2341 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2562 -0.6082 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2562 -0.6082 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8090 -0.5806 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8090 -0.5806 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7335 -1.2705 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7335 -1.2705 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3339 0.5904 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3339 0.5904 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9344 1.3122 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9344 1.3122 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3597 2.0191 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3597 2.0191 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.1846 2.0042 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.1846 2.0042 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.5841 1.2824 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.5841 1.2824 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.1588 0.5755 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.1588 0.5755 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.5577 -0.1454 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.5577 -0.1454 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.4079 1.2675 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.4079 1.2675 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.6094 2.7101 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.6094 2.7101 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1106 1.3271 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1106 1.3271 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7117 2.0480 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7117 2.0480 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6858 0.6212 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6858 0.6212 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 2 7 1 0 0 0 0 | + | 2 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 11 1 0 0 0 0 | + | 10 11 1 0 0 0 0 |
| − | 11 12 1 0 0 0 0 | + | 11 12 1 0 0 0 0 |
| − | 12 7 2 0 0 0 0 | + | 12 7 2 0 0 0 0 |
| − | 9 13 1 0 0 0 0 | + | 9 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 10 1 0 0 0 0 | + | 16 10 1 0 0 0 0 |
| − | 14 18 1 0 0 0 0 | + | 14 18 1 0 0 0 0 |
| − | 11 17 2 0 0 0 0 | + | 11 17 2 0 0 0 0 |
| − | 12 19 1 0 0 0 0 | + | 12 19 1 0 0 0 0 |
| − | 16 20 1 0 0 0 0 | + | 16 20 1 0 0 0 0 |
| − | 5 21 1 0 0 0 0 | + | 5 21 1 0 0 0 0 |
| − | 4 22 1 0 0 0 0 | + | 4 22 1 0 0 0 0 |
| − | 6 23 1 0 0 0 0 | + | 6 23 1 0 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 25 26 1 1 0 0 0 | + | 25 26 1 1 0 0 0 |
| − | 27 26 1 1 0 0 0 | + | 27 26 1 1 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 29 24 1 0 0 0 0 | + | 29 24 1 0 0 0 0 |
| − | 29 30 1 0 0 0 0 | + | 29 30 1 0 0 0 0 |
| − | 24 31 1 0 0 0 0 | + | 24 31 1 0 0 0 0 |
| − | 25 32 1 0 0 0 0 | + | 25 32 1 0 0 0 0 |
| − | 26 33 1 0 0 0 0 | + | 26 33 1 0 0 0 0 |
| − | 27 18 1 0 0 0 0 | + | 27 18 1 0 0 0 0 |
| − | 34 35 2 0 0 0 0 | + | 34 35 2 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 2 0 0 0 0 | + | 36 37 2 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 2 0 0 0 0 | + | 38 39 2 0 0 0 0 |
| − | 39 34 1 0 0 0 0 | + | 39 34 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 38 41 1 0 0 0 0 | + | 38 41 1 0 0 0 0 |
| − | 37 42 1 0 0 0 0 | + | 37 42 1 0 0 0 0 |
| − | 35 43 1 0 0 0 0 | + | 35 43 1 0 0 0 0 |
| − | 43 44 2 0 0 0 0 | + | 43 44 2 0 0 0 0 |
| − | 43 45 1 0 0 0 0 | + | 43 45 1 0 0 0 0 |
| − | 45 30 1 0 0 0 0 | + | 45 30 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FAGGS0032 | + | ID FL5FAGGS0032 |
| − | KNApSAcK_ID C00013974 | + | KNApSAcK_ID C00013974 |
| − | NAME Myricetin 7-(6''-galloylglucoside);3,5-Dihydroxy-7-[[6-O-(3,4,5-trihydroxybenzoyl)-beta-D-glucopyranosyl]oxy]-2-(3,4,5-trihydroxyphenyl)-4H-1-benzopyran-4-one | + | NAME Myricetin 7-(6''-galloylglucoside);3,5-Dihydroxy-7-[[6-O-(3,4,5-trihydroxybenzoyl)-beta-D-glucopyranosyl]oxy]-2-(3,4,5-trihydroxyphenyl)-4H-1-benzopyran-4-one |
| − | CAS_RN 231289-26-2 | + | CAS_RN 231289-26-2 |
| − | FORMULA C28H24O17 | + | FORMULA C28H24O17 |
| − | EXACTMASS 632.101349342 | + | EXACTMASS 632.101349342 |
| − | AVERAGEMASS 632.47996 | + | AVERAGEMASS 632.47996 |
| − | SMILES C(C(O)1)(O)C(C(Oc(c5)cc(O)c(c35)C(=O)C(=C(c(c4)cc(O)c(c4O)O)O3)O)OC1COC(=O)c(c2)cc(O)c(c(O)2)O)O | + | SMILES C(C(O)1)(O)C(C(Oc(c5)cc(O)c(c35)C(=O)C(=C(c(c4)cc(O)c(c4O)O)O3)O)OC1COC(=O)c(c2)cc(O)c(c(O)2)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
45 49 0 0 0 0 0 0 0 0999 V2000
4.9789 -0.7657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2644 -0.3532 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2644 0.4718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.9789 0.8843 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.6934 0.4718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.6934 -0.3532 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5500 -0.7657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8355 -0.3532 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1210 -0.7657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1210 -1.5907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8355 -2.0032 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5500 -1.5907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4066 -0.3532 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6921 -0.7657 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6921 -1.5907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4066 -2.0032 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8355 -2.7101 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0224 -0.3532 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.2644 -2.0032 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4066 -2.6942 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
6.4079 0.8843 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.9789 1.7093 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
6.3713 -0.7446 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7730 -0.1250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3603 -0.8397 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5620 -0.6305 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7685 -0.8574 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1811 -0.1426 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9795 -0.3517 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1365 0.2341 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2562 -0.6082 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8090 -0.5806 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7335 -1.2705 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.3339 0.5904 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9344 1.3122 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3597 2.0191 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.1846 2.0042 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.5841 1.2824 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.1588 0.5755 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.5577 -0.1454 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-6.4079 1.2675 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.6094 2.7101 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1106 1.3271 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7117 2.0480 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6858 0.6212 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
2 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 11 1 0 0 0 0
11 12 1 0 0 0 0
12 7 2 0 0 0 0
9 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 10 1 0 0 0 0
14 18 1 0 0 0 0
11 17 2 0 0 0 0
12 19 1 0 0 0 0
16 20 1 0 0 0 0
5 21 1 0 0 0 0
4 22 1 0 0 0 0
6 23 1 0 0 0 0
24 25 1 1 0 0 0
25 26 1 1 0 0 0
27 26 1 1 0 0 0
27 28 1 0 0 0 0
28 29 1 0 0 0 0
29 24 1 0 0 0 0
29 30 1 0 0 0 0
24 31 1 0 0 0 0
25 32 1 0 0 0 0
26 33 1 0 0 0 0
27 18 1 0 0 0 0
34 35 2 0 0 0 0
35 36 1 0 0 0 0
36 37 2 0 0 0 0
37 38 1 0 0 0 0
38 39 2 0 0 0 0
39 34 1 0 0 0 0
39 40 1 0 0 0 0
38 41 1 0 0 0 0
37 42 1 0 0 0 0
35 43 1 0 0 0 0
43 44 2 0 0 0 0
43 45 1 0 0 0 0
45 30 1 0 0 0 0
S SKP 8
ID FL5FAGGS0032
KNApSAcK_ID C00013974
NAME Myricetin 7-(6''-galloylglucoside);3,5-Dihydroxy-7-[[6-O-(3,4,5-trihydroxybenzoyl)-beta-D-glucopyranosyl]oxy]-2-(3,4,5-trihydroxyphenyl)-4H-1-benzopyran-4-one
CAS_RN 231289-26-2
FORMULA C28H24O17
EXACTMASS 632.101349342
AVERAGEMASS 632.47996
SMILES C(C(O)1)(O)C(C(Oc(c5)cc(O)c(c35)C(=O)C(=C(c(c4)cc(O)c(c4O)O)O3)O)OC1COC(=O)c(c2)cc(O)c(c(O)2)O)O
M END