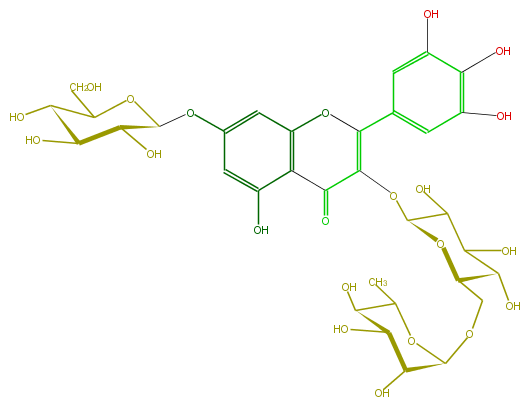
Mol:FL5FAGGL0008
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 55 60 0 0 0 0 0 0 0 0999 V2000 | + | 55 60 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.7419 1.4721 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7419 1.4721 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7419 0.6627 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7419 0.6627 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0408 0.2578 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0408 0.2578 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6602 0.6626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6602 0.6626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6601 1.4721 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6601 1.4721 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0409 1.8769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0409 1.8769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3614 0.2579 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3614 0.2579 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0623 0.6626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0623 0.6626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0624 1.4721 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0624 1.4721 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3613 1.8768 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3613 1.8768 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3616 -0.3732 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3616 -0.3732 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7633 1.8767 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7633 1.8767 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4778 1.4642 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4778 1.4642 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1922 1.8768 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1922 1.8768 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1922 2.7018 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1922 2.7018 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4778 3.1143 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4778 3.1143 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7632 2.7019 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7632 2.7019 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4427 1.8768 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4427 1.8768 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7873 0.1504 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7873 0.1504 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0407 -0.5513 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0407 -0.5513 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8116 -2.0830 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8116 -2.0830 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9784 -2.2299 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9784 -2.2299 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6795 -2.7037 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6795 -2.7037 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0275 -3.4802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0275 -3.4802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8608 -3.3333 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8608 -3.3333 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1598 -2.8594 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1598 -2.8594 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3288 -1.6294 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3288 -1.6294 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8969 -0.2190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8969 -0.2190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0636 -0.3659 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0636 -0.3659 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7647 -0.8397 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7647 -0.8397 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1128 -1.6162 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1128 -1.6162 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9460 -1.4693 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9460 -1.4693 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2450 -0.9954 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2450 -0.9954 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3144 0.3081 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3144 0.3081 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1124 -0.9836 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1124 -0.9836 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1910 -2.1376 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1910 -2.1376 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6127 -2.0262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6127 -2.0262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3679 -2.7143 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3679 -2.7143 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7823 -1.7392 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7823 -1.7392 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7708 -2.6132 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7708 -2.6132 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4806 -3.9392 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4806 -3.9392 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9911 3.1631 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9911 3.1631 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4777 3.9392 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4777 3.9392 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3551 2.0157 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3551 2.0157 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7612 1.2316 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7612 1.2316 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9060 1.5643 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9060 1.5643 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0806 1.5731 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0806 1.5731 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6804 2.1730 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6804 2.1730 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4285 1.7781 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4285 1.7781 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9697 1.7872 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9697 1.7872 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6239 1.3090 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6239 1.3090 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2687 1.0277 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2687 1.0277 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0140 1.8048 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0140 1.8048 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9866 2.4189 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9866 2.4189 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.1910 2.7898 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.1910 2.7898 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 8 19 1 0 0 0 0 | + | 8 19 1 0 0 0 0 |
| − | 3 20 1 0 0 0 0 | + | 3 20 1 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 29 30 1 1 0 0 0 | + | 29 30 1 1 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 32 31 1 1 0 0 0 | + | 32 31 1 1 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 28 1 0 0 0 0 | + | 33 28 1 0 0 0 0 |
| − | 28 34 1 0 0 0 0 | + | 28 34 1 0 0 0 0 |
| − | 33 35 1 0 0 0 0 | + | 33 35 1 0 0 0 0 |
| − | 32 36 1 0 0 0 0 | + | 32 36 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 25 38 1 0 0 0 0 | + | 25 38 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 22 39 1 0 0 0 0 | + | 22 39 1 0 0 0 0 |
| − | 23 40 1 0 0 0 0 | + | 23 40 1 0 0 0 0 |
| − | 24 41 1 0 0 0 0 | + | 24 41 1 0 0 0 0 |
| − | 29 19 1 0 0 0 0 | + | 29 19 1 0 0 0 0 |
| − | 42 15 1 0 0 0 0 | + | 42 15 1 0 0 0 0 |
| − | 16 43 1 0 0 0 0 | + | 16 43 1 0 0 0 0 |
| − | 44 45 1 1 0 0 0 | + | 44 45 1 1 0 0 0 |
| − | 45 46 1 1 0 0 0 | + | 45 46 1 1 0 0 0 |
| − | 47 46 1 1 0 0 0 | + | 47 46 1 1 0 0 0 |
| − | 47 48 1 0 0 0 0 | + | 47 48 1 0 0 0 0 |
| − | 48 49 1 0 0 0 0 | + | 48 49 1 0 0 0 0 |
| − | 49 44 1 0 0 0 0 | + | 49 44 1 0 0 0 0 |
| − | 44 50 1 0 0 0 0 | + | 44 50 1 0 0 0 0 |
| − | 45 51 1 0 0 0 0 | + | 45 51 1 0 0 0 0 |
| − | 46 52 1 0 0 0 0 | + | 46 52 1 0 0 0 0 |
| − | 47 18 1 0 0 0 0 | + | 47 18 1 0 0 0 0 |
| − | 14 53 1 0 0 0 0 | + | 14 53 1 0 0 0 0 |
| − | 54 55 1 0 0 0 0 | + | 54 55 1 0 0 0 0 |
| − | 49 54 1 0 0 0 0 | + | 49 54 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 54 55 | + | M SAL 1 2 54 55 |
| − | M SBL 1 1 60 | + | M SBL 1 1 60 |
| − | M SMT 1 ^ CH2OH | + | M SMT 1 ^ CH2OH |
| − | M SBV 1 60 0.5581 -0.6407 | + | M SBV 1 60 0.5581 -0.6407 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FAGGL0008 | + | ID FL5FAGGL0008 |
| − | FORMULA C33H40O22 | + | FORMULA C33H40O22 |
| − | EXACTMASS 788.201122964 | + | EXACTMASS 788.201122964 |
| − | AVERAGEMASS 788.6575 | + | AVERAGEMASS 788.6575 |
| − | SMILES C(C6O)(O)C(OC(C6O)C)OCC(C(O)1)OC(OC(C(=O)5)=C(Oc(c54)cc(cc(O)4)OC(O3)C(O)C(C(O)C3CO)O)c(c2)cc(O)c(c(O)2)O)C(C1O)O | + | SMILES C(C6O)(O)C(OC(C6O)C)OCC(C(O)1)OC(OC(C(=O)5)=C(Oc(c54)cc(cc(O)4)OC(O3)C(O)C(C(O)C3CO)O)c(c2)cc(O)c(c(O)2)O)C(C1O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
55 60 0 0 0 0 0 0 0 0999 V2000
-0.7419 1.4721 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7419 0.6627 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0408 0.2578 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6602 0.6626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6601 1.4721 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0409 1.8769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3614 0.2579 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0623 0.6626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0624 1.4721 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3613 1.8768 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3616 -0.3732 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7633 1.8767 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4778 1.4642 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1922 1.8768 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1922 2.7018 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4778 3.1143 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7632 2.7019 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4427 1.8768 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7873 0.1504 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0407 -0.5513 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8116 -2.0830 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9784 -2.2299 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6795 -2.7037 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0275 -3.4802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8608 -3.3333 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1598 -2.8594 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3288 -1.6294 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8969 -0.2190 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0636 -0.3659 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7647 -0.8397 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1128 -1.6162 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.9460 -1.4693 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2450 -0.9954 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3144 0.3081 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.1124 -0.9836 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.1910 -2.1376 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.6127 -2.0262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3679 -2.7143 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7823 -1.7392 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7708 -2.6132 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4806 -3.9392 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.9911 3.1631 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4777 3.9392 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.3551 2.0157 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7612 1.2316 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9060 1.5643 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0806 1.5731 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6804 2.1730 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4285 1.7781 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.9697 1.7872 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.6239 1.3090 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2687 1.0277 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.0140 1.8048 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9866 2.4189 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.1910 2.7898 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
8 19 1 0 0 0 0
3 20 1 0 0 0 0
21 22 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
28 29 1 0 0 0 0
29 30 1 1 0 0 0
30 31 1 1 0 0 0
32 31 1 1 0 0 0
32 33 1 0 0 0 0
33 28 1 0 0 0 0
28 34 1 0 0 0 0
33 35 1 0 0 0 0
32 36 1 0 0 0 0
31 37 1 0 0 0 0
25 38 1 0 0 0 0
37 38 1 0 0 0 0
22 39 1 0 0 0 0
23 40 1 0 0 0 0
24 41 1 0 0 0 0
29 19 1 0 0 0 0
42 15 1 0 0 0 0
16 43 1 0 0 0 0
44 45 1 1 0 0 0
45 46 1 1 0 0 0
47 46 1 1 0 0 0
47 48 1 0 0 0 0
48 49 1 0 0 0 0
49 44 1 0 0 0 0
44 50 1 0 0 0 0
45 51 1 0 0 0 0
46 52 1 0 0 0 0
47 18 1 0 0 0 0
14 53 1 0 0 0 0
54 55 1 0 0 0 0
49 54 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 54 55
M SBL 1 1 60
M SMT 1 ^ CH2OH
M SBV 1 60 0.5581 -0.6407
S SKP 5
ID FL5FAGGL0008
FORMULA C33H40O22
EXACTMASS 788.201122964
AVERAGEMASS 788.6575
SMILES C(C6O)(O)C(OC(C6O)C)OCC(C(O)1)OC(OC(C(=O)5)=C(Oc(c54)cc(cc(O)4)OC(O3)C(O)C(C(O)C3CO)O)c(c2)cc(O)c(c(O)2)O)C(C1O)O
M END