Mol:FL5FAGGA0003
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 45 49 0 0 0 0 0 0 0 0999 V2000 | + | 45 49 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.5772 -1.0723 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5772 -1.0723 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5772 -1.7147 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5772 -1.7147 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0209 -2.0359 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0209 -2.0359 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4646 -1.7147 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4646 -1.7147 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4646 -1.0723 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4646 -1.0723 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0209 -0.7512 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0209 -0.7512 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9083 -2.0359 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9083 -2.0359 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3520 -1.7147 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3520 -1.7147 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3520 -1.0723 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3520 -1.0723 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9083 -0.7512 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9083 -0.7512 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9083 -2.5367 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9083 -2.5367 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3484 -0.6275 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3484 -0.6275 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9154 -0.9549 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9154 -0.9549 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4824 -0.6275 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4824 -0.6275 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4824 0.0272 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4824 0.0272 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9154 0.3545 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9154 0.3545 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3484 0.0272 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3484 0.0272 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0209 -2.6780 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0209 -2.6780 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0892 0.3775 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0892 0.3775 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2417 -0.6887 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2417 -0.6887 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0920 -2.1587 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0920 -2.1587 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8713 0.9695 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8713 0.9695 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0492 -0.9548 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0492 -0.9548 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3040 -1.7228 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 1.3040 -1.7228 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 1.0032 -2.2439 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.0032 -2.2439 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.5817 -2.0785 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5817 -2.0785 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1636 -2.2439 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.1636 -2.2439 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.4645 -1.7228 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.4645 -1.7228 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.8860 -1.8881 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 1.8860 -1.8881 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 0.7453 -1.8725 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7453 -1.8725 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7231 -1.4405 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7231 -1.4405 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4645 -1.1537 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4645 -1.1537 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3008 2.6780 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -0.3008 2.6780 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -0.5395 2.1116 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -0.5395 2.1116 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -0.0922 1.7526 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -0.0922 1.7526 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 0.1705 1.3088 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 0.1705 1.3088 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 0.3077 1.8208 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3077 1.8208 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1398 2.1023 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -0.1398 2.1023 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -0.5722 3.6405 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5722 3.6405 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8532 2.5986 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8532 2.5986 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3843 1.2303 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3843 1.2303 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8602 2.1023 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8602 2.1023 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3602 2.9683 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3602 2.9683 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3341 -2.5654 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3341 -2.5654 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3341 -3.3904 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3341 -3.3904 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 1 20 1 0 0 0 0 | + | 1 20 1 0 0 0 0 |
| − | 21 8 1 0 0 0 0 | + | 21 8 1 0 0 0 0 |
| − | 16 22 1 0 0 0 0 | + | 16 22 1 0 0 0 0 |
| − | 14 23 1 0 0 0 0 | + | 14 23 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 1 0 0 0 | + | 25 26 1 1 0 0 0 |
| − | 26 27 1 1 0 0 0 | + | 26 27 1 1 0 0 0 |
| − | 28 27 1 1 0 0 0 | + | 28 27 1 1 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 29 24 1 0 0 0 0 | + | 29 24 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 29 31 1 0 0 0 0 | + | 29 31 1 0 0 0 0 |
| − | 28 32 1 0 0 0 0 | + | 28 32 1 0 0 0 0 |
| − | 25 21 1 0 0 0 0 | + | 25 21 1 0 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 34 35 1 1 0 0 0 | + | 34 35 1 1 0 0 0 |
| − | 36 35 1 1 0 0 0 | + | 36 35 1 1 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 33 1 0 0 0 0 | + | 38 33 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 34 40 1 0 0 0 0 | + | 34 40 1 0 0 0 0 |
| − | 35 41 1 0 0 0 0 | + | 35 41 1 0 0 0 0 |
| − | 36 22 1 0 0 0 0 | + | 36 22 1 0 0 0 0 |
| − | 38 42 1 0 0 0 0 | + | 38 42 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 27 44 1 0 0 0 0 | + | 27 44 1 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 44 45 | + | M SAL 2 2 44 45 |
| − | M SBL 2 1 48 | + | M SBL 2 1 48 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SVB 2 48 2.5273 -2.257 | + | M SVB 2 48 2.5273 -2.257 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 42 43 | + | M SAL 1 2 42 43 |
| − | M SBL 1 1 46 | + | M SBL 1 1 46 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SVB 1 46 0.2427 2.4886 | + | M SVB 1 46 0.2427 2.4886 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FAGGA0003 | + | ID FL5FAGGA0003 |
| − | KNApSAcK_ID C00005743 | + | KNApSAcK_ID C00005743 |
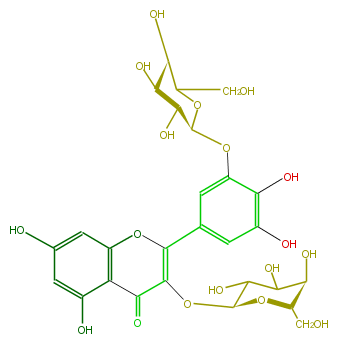
| − | NAME Myricetin 3,3'-digalactoside | + | NAME Myricetin 3,3'-digalactoside |
| − | CAS_RN 28454-81-1 | + | CAS_RN 28454-81-1 |
| − | FORMULA C27H30O18 | + | FORMULA C27H30O18 |
| − | EXACTMASS 642.143214156 | + | EXACTMASS 642.143214156 |
| − | AVERAGEMASS 642.5163 | + | AVERAGEMASS 642.5163 |
| − | SMILES O([C@@H](Oc(c(O)5)cc(cc(O)5)C(O2)=C(O[C@@H](C(O)4)O[C@H](CO)[C@@H](C(O)4)O)C(=O)c(c3O)c2cc(c3)O)1)C(CO)[C@@H]([C@@H]([C@@H]1O)O)O | + | SMILES O([C@@H](Oc(c(O)5)cc(cc(O)5)C(O2)=C(O[C@@H](C(O)4)O[C@H](CO)[C@@H](C(O)4)O)C(=O)c(c3O)c2cc(c3)O)1)C(CO)[C@@H]([C@@H]([C@@H]1O)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
45 49 0 0 0 0 0 0 0 0999 V2000
-2.5772 -1.0723 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5772 -1.7147 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0209 -2.0359 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4646 -1.7147 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4646 -1.0723 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0209 -0.7512 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9083 -2.0359 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3520 -1.7147 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3520 -1.0723 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9083 -0.7512 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9083 -2.5367 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3484 -0.6275 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9154 -0.9549 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4824 -0.6275 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4824 0.0272 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9154 0.3545 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3484 0.0272 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0209 -2.6780 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0892 0.3775 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2417 -0.6887 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0920 -2.1587 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8713 0.9695 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0492 -0.9548 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3040 -1.7228 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
1.0032 -2.2439 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.5817 -2.0785 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1636 -2.2439 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.4645 -1.7228 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.8860 -1.8881 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
0.7453 -1.8725 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7231 -1.4405 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4645 -1.1537 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.3008 2.6780 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-0.5395 2.1116 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.0922 1.7526 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.1705 1.3088 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.3077 1.8208 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1398 2.1023 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-0.5722 3.6405 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8532 2.5986 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.3843 1.2303 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8602 2.1023 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3602 2.9683 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3341 -2.5654 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3341 -3.3904 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
4 3 1 0 0 0 0
1 20 1 0 0 0 0
21 8 1 0 0 0 0
16 22 1 0 0 0 0
14 23 1 0 0 0 0
24 25 1 0 0 0 0
25 26 1 1 0 0 0
26 27 1 1 0 0 0
28 27 1 1 0 0 0
28 29 1 0 0 0 0
29 24 1 0 0 0 0
24 30 1 0 0 0 0
29 31 1 0 0 0 0
28 32 1 0 0 0 0
25 21 1 0 0 0 0
33 34 1 1 0 0 0
34 35 1 1 0 0 0
36 35 1 1 0 0 0
36 37 1 0 0 0 0
37 38 1 0 0 0 0
38 33 1 0 0 0 0
33 39 1 0 0 0 0
34 40 1 0 0 0 0
35 41 1 0 0 0 0
36 22 1 0 0 0 0
38 42 1 0 0 0 0
42 43 1 0 0 0 0
27 44 1 0 0 0 0
44 45 1 0 0 0 0
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 44 45
M SBL 2 1 48
M SMT 2 CH2OH
M SVB 2 48 2.5273 -2.257
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 42 43
M SBL 1 1 46
M SMT 1 CH2OH
M SVB 1 46 0.2427 2.4886
S SKP 8
ID FL5FAGGA0003
KNApSAcK_ID C00005743
NAME Myricetin 3,3'-digalactoside
CAS_RN 28454-81-1
FORMULA C27H30O18
EXACTMASS 642.143214156
AVERAGEMASS 642.5163
SMILES O([C@@H](Oc(c(O)5)cc(cc(O)5)C(O2)=C(O[C@@H](C(O)4)O[C@H](CO)[C@@H](C(O)4)O)C(=O)c(c3O)c2cc(c3)O)1)C(CO)[C@@H]([C@@H]([C@@H]1O)O)O
M END