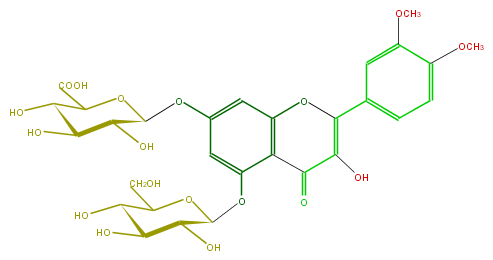
Mol:FL5FAFGS0002
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 47 51 0 0 0 0 0 0 0 0999 V2000 | + | 47 51 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.3161 -0.3259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3161 -0.3259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3161 -1.1509 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3161 -1.1509 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6017 -1.5634 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6017 -1.5634 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1128 -1.1509 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1128 -1.1509 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1128 -0.3259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1128 -0.3259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6017 0.0866 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6017 0.0866 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8272 -1.5634 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8272 -1.5634 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5416 -1.1509 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5416 -1.1509 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5416 -0.3259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5416 -0.3259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8272 0.0866 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8272 0.0866 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8272 -2.2065 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8272 -2.2065 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2558 0.0864 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2558 0.0864 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9838 -0.3340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9838 -0.3340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7120 0.0864 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7120 0.0864 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7120 0.9272 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7120 0.9272 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9838 1.3476 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9838 1.3476 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2558 0.9272 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2558 0.9272 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0302 0.0864 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0302 0.0864 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0477 -1.6503 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0477 -1.6503 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6001 -2.2035 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6001 -2.2035 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8964 0.0123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8964 0.0123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3482 -0.7115 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3482 -0.7115 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5586 -0.4045 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5586 -0.4045 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7968 -0.3962 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7968 -0.3962 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3503 0.1575 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3503 0.1575 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1568 -0.1321 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1568 -0.1321 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.6620 -0.1748 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.6620 -0.1748 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.2356 -0.6104 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.2356 -0.6104 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8573 -0.9383 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8573 -0.9383 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3577 -2.2892 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3577 -2.2892 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8095 -3.0128 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8095 -3.0128 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0199 -2.7058 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0199 -2.7058 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2581 -2.6977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2581 -2.6977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8116 -2.1439 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8116 -2.1439 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6181 -2.4335 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6181 -2.4335 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1645 -2.5175 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1645 -2.5175 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6628 -2.9115 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6628 -2.9115 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3186 -3.2398 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3186 -3.2398 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4160 1.3336 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4160 1.3336 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.7003 1.3336 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.7003 1.3336 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2130 -1.7885 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2130 -1.7885 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3999 -0.7944 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3999 -0.7944 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9838 2.0874 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9838 2.0874 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5507 3.2398 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5507 3.2398 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8324 0.4585 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8324 0.4585 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.7003 1.0662 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.7003 1.0662 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8687 0.9079 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8687 0.9079 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 8 19 1 0 0 0 0 | + | 8 19 1 0 0 0 0 |
| − | 3 20 1 0 0 0 0 | + | 3 20 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 18 1 0 0 0 0 | + | 24 18 1 0 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 33 32 1 1 0 0 0 | + | 33 32 1 1 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 30 1 0 0 0 0 | + | 35 30 1 0 0 0 0 |
| − | 30 36 1 0 0 0 0 | + | 30 36 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 20 1 0 0 0 0 | + | 33 20 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 15 39 1 0 0 0 0 | + | 15 39 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 35 41 1 0 0 0 0 | + | 35 41 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 16 43 1 0 0 0 0 | + | 16 43 1 0 0 0 0 |
| − | 45 46 2 0 0 0 0 | + | 45 46 2 0 0 0 0 |
| − | 45 47 1 0 0 0 0 | + | 45 47 1 0 0 0 0 |
| − | 26 45 1 0 0 0 0 | + | 26 45 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 39 40 | + | M SAL 1 2 39 40 |
| − | M SBL 1 1 44 | + | M SBL 1 1 44 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 44 -0.7040 -0.4065 | + | M SBV 1 44 -0.7040 -0.4065 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 41 42 | + | M SAL 2 2 41 42 |
| − | M SBL 2 1 46 | + | M SBL 2 1 46 |
| − | M SMT 2 ^ CH2OH | + | M SMT 2 ^ CH2OH |
| − | M SBV 2 46 0.5948 -0.6450 | + | M SBV 2 46 0.5948 -0.6450 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 43 44 | + | M SAL 3 2 43 44 |
| − | M SBL 3 1 48 | + | M SBL 3 1 48 |
| − | M SMT 3 OCH3 | + | M SMT 3 OCH3 |
| − | M SBV 3 48 0.0000 -0.7398 | + | M SBV 3 48 0.0000 -0.7398 |
| − | M STY 1 4 SUP | + | M STY 1 4 SUP |
| − | M SLB 1 4 4 | + | M SLB 1 4 4 |
| − | M SAL 4 3 45 46 47 | + | M SAL 4 3 45 46 47 |
| − | M SBL 4 1 51 | + | M SBL 4 1 51 |
| − | M SMT 4 ^ COOH | + | M SMT 4 ^ COOH |
| − | M SBV 4 51 0.6756 -0.5906 | + | M SBV 4 51 0.6756 -0.5906 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FAFGS0002 | + | ID FL5FAFGS0002 |
| − | FORMULA C29H32O18 | + | FORMULA C29H32O18 |
| − | EXACTMASS 668.1588642199999 | + | EXACTMASS 668.1588642199999 |
| − | AVERAGEMASS 668.55358 | + | AVERAGEMASS 668.55358 |
| − | SMILES c(c(OC(O5)C(C(O)C(O)C5CO)O)2)(C(=O)3)c(OC(c(c4)ccc(c4OC)OC)=C3O)cc(c2)OC(C(O)1)OC(C(O)=O)C(O)C1O | + | SMILES c(c(OC(O5)C(C(O)C(O)C5CO)O)2)(C(=O)3)c(OC(c(c4)ccc(c4OC)OC)=C3O)cc(c2)OC(C(O)1)OC(C(O)=O)C(O)C1O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
47 51 0 0 0 0 0 0 0 0999 V2000
-1.3161 -0.3259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3161 -1.1509 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6017 -1.5634 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1128 -1.1509 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1128 -0.3259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6017 0.0866 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8272 -1.5634 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5416 -1.1509 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5416 -0.3259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8272 0.0866 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8272 -2.2065 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2558 0.0864 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9838 -0.3340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7120 0.0864 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7120 0.9272 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9838 1.3476 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2558 0.9272 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0302 0.0864 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0477 -1.6503 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6001 -2.2035 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.8964 0.0123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3482 -0.7115 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5586 -0.4045 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7968 -0.3962 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3503 0.1575 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1568 -0.1321 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.6620 -0.1748 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.2356 -0.6104 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8573 -0.9383 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3577 -2.2892 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8095 -3.0128 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0199 -2.7058 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2581 -2.6977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8116 -2.1439 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6181 -2.4335 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1645 -2.5175 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6628 -2.9115 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3186 -3.2398 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.4160 1.3336 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.7003 1.3336 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2130 -1.7885 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3999 -0.7944 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9838 2.0874 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5507 3.2398 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.8324 0.4585 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.7003 1.0662 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.8687 0.9079 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
8 19 1 0 0 0 0
3 20 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
24 18 1 0 0 0 0
30 31 1 1 0 0 0
31 32 1 1 0 0 0
33 32 1 1 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 30 1 0 0 0 0
30 36 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 20 1 0 0 0 0
39 40 1 0 0 0 0
15 39 1 0 0 0 0
41 42 1 0 0 0 0
35 41 1 0 0 0 0
43 44 1 0 0 0 0
16 43 1 0 0 0 0
45 46 2 0 0 0 0
45 47 1 0 0 0 0
26 45 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 39 40
M SBL 1 1 44
M SMT 1 OCH3
M SBV 1 44 -0.7040 -0.4065
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 41 42
M SBL 2 1 46
M SMT 2 ^ CH2OH
M SBV 2 46 0.5948 -0.6450
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 43 44
M SBL 3 1 48
M SMT 3 OCH3
M SBV 3 48 0.0000 -0.7398
M STY 1 4 SUP
M SLB 1 4 4
M SAL 4 3 45 46 47
M SBL 4 1 51
M SMT 4 ^ COOH
M SBV 4 51 0.6756 -0.5906
S SKP 5
ID FL5FAFGS0002
FORMULA C29H32O18
EXACTMASS 668.1588642199999
AVERAGEMASS 668.55358
SMILES c(c(OC(O5)C(C(O)C(O)C5CO)O)2)(C(=O)3)c(OC(c(c4)ccc(c4OC)OC)=C3O)cc(c2)OC(C(O)1)OC(C(O)=O)C(O)C1O
M END