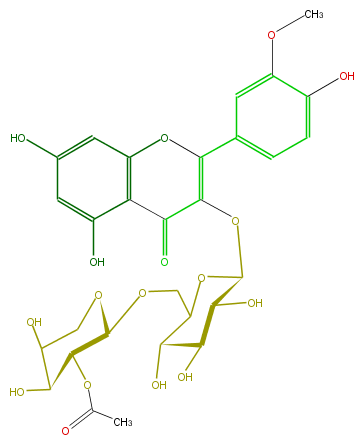
Mol:FL5FADGS0030
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 46 50 0 0 0 0 0 0 0 0999 V2000 | + | 46 50 0 0 0 0 0 0 0 0999 V2000 |
| − | 1.7975 1.2736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7975 1.2736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0831 1.6861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0831 1.6861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0831 2.5110 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0831 2.5110 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7975 2.9235 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7975 2.9235 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5119 2.5110 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5119 2.5110 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5119 1.6861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5119 1.6861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3686 1.2736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3686 1.2736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3459 1.6861 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3459 1.6861 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0603 1.2736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0603 1.2736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0603 0.4486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0603 0.4486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3459 0.0361 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3459 0.0361 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3686 0.4486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3686 0.4486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7748 1.6861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7748 1.6861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4892 1.2736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4892 1.2736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4892 0.4486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4892 0.4486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7748 0.0361 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7748 0.0361 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3459 -0.7889 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3459 -0.7889 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2036 1.6861 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2036 1.6861 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0831 0.0361 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0831 0.0361 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7748 -0.7889 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7748 -0.7889 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2264 2.9235 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2264 2.9235 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7975 3.7485 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7975 3.7485 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5292 4.1710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5292 4.1710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4404 -2.4625 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4404 -2.4625 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3847 -2.4721 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3847 -2.4721 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6119 -1.6787 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6119 -1.6787 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2117 -1.1119 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2117 -1.1119 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3866 -1.1023 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3866 -1.1023 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1592 -1.8956 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1592 -1.8956 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1013 -1.3816 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1013 -1.3816 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7898 -1.4124 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7898 -1.4124 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5222 -3.2470 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5222 -3.2470 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0057 -3.0803 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0057 -3.0803 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3890 -1.6096 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3890 -1.6096 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8251 -2.5484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8251 -2.5484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6392 -3.3524 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6392 -3.3524 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2138 -2.6452 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2138 -2.6452 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4874 -2.2536 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4874 -2.2536 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6732 -1.4495 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6732 -1.4495 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0987 -2.1566 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0987 -2.1566 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0355 -1.9820 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0355 -1.9820 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2264 -3.3881 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2264 -3.3881 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8903 -3.2531 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8903 -3.2531 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8059 -3.7703 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8059 -3.7703 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3287 -4.1710 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3287 -4.1710 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2975 -3.9814 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2975 -3.9814 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 2 7 1 0 0 0 0 | + | 2 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 11 1 0 0 0 0 | + | 10 11 1 0 0 0 0 |
| − | 11 12 1 0 0 0 0 | + | 11 12 1 0 0 0 0 |
| − | 12 7 2 0 0 0 0 | + | 12 7 2 0 0 0 0 |
| − | 9 13 1 0 0 0 0 | + | 9 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 10 1 0 0 0 0 | + | 16 10 1 0 0 0 0 |
| − | 14 18 1 0 0 0 0 | + | 14 18 1 0 0 0 0 |
| − | 11 17 2 0 0 0 0 | + | 11 17 2 0 0 0 0 |
| − | 12 19 1 0 0 0 0 | + | 12 19 1 0 0 0 0 |
| − | 16 20 1 0 0 0 0 | + | 16 20 1 0 0 0 0 |
| − | 5 21 1 0 0 0 0 | + | 5 21 1 0 0 0 0 |
| − | 4 22 1 0 0 0 0 | + | 4 22 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 25 26 1 1 0 0 0 | + | 25 26 1 1 0 0 0 |
| − | 27 26 1 1 0 0 0 | + | 27 26 1 1 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 29 24 1 0 0 0 0 | + | 29 24 1 0 0 0 0 |
| − | 29 30 1 0 0 0 0 | + | 29 30 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 24 32 1 0 0 0 0 | + | 24 32 1 0 0 0 0 |
| − | 25 33 1 0 0 0 0 | + | 25 33 1 0 0 0 0 |
| − | 26 34 1 0 0 0 0 | + | 26 34 1 0 0 0 0 |
| − | 27 19 1 0 0 0 0 | + | 27 19 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 1 1 0 0 0 | + | 36 37 1 1 0 0 0 |
| − | 38 37 1 1 0 0 0 | + | 38 37 1 1 0 0 0 |
| − | 39 38 1 1 0 0 0 | + | 39 38 1 1 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 40 35 1 0 0 0 0 | + | 40 35 1 0 0 0 0 |
| − | 35 41 1 0 0 0 0 | + | 35 41 1 0 0 0 0 |
| − | 36 42 1 0 0 0 0 | + | 36 42 1 0 0 0 0 |
| − | 37 43 1 0 0 0 0 | + | 37 43 1 0 0 0 0 |
| − | 38 31 1 0 0 0 0 | + | 38 31 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 44 45 2 0 0 0 0 | + | 44 45 2 0 0 0 0 |
| − | 44 46 1 0 0 0 0 | + | 44 46 1 0 0 0 0 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FADGS0030 | + | ID FL5FADGS0030 |
| − | FORMULA C29H32O17 | + | FORMULA C29H32O17 |
| − | EXACTMASS 652.163949598 | + | EXACTMASS 652.163949598 |
| − | AVERAGEMASS 652.55418 | + | AVERAGEMASS 652.55418 |
| − | SMILES C(C(O)1)(O)COC(OCC(O2)C(O)C(C(O)C2OC(=C4c(c5)cc(OC)c(O)c5)C(=O)c(c(O4)3)c(cc(O)c3)O)O)C1OC(C)=O | + | SMILES C(C(O)1)(O)COC(OCC(O2)C(O)C(C(O)C2OC(=C4c(c5)cc(OC)c(O)c5)C(=O)c(c(O4)3)c(cc(O)c3)O)O)C1OC(C)=O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
46 50 0 0 0 0 0 0 0 0999 V2000
1.7975 1.2736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0831 1.6861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0831 2.5110 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7975 2.9235 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5119 2.5110 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5119 1.6861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3686 1.2736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3459 1.6861 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0603 1.2736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0603 0.4486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3459 0.0361 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3686 0.4486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7748 1.6861 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4892 1.2736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4892 0.4486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7748 0.0361 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3459 -0.7889 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2036 1.6861 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0831 0.0361 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7748 -0.7889 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2264 2.9235 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7975 3.7485 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5292 4.1710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4404 -2.4625 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3847 -2.4721 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6119 -1.6787 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2117 -1.1119 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3866 -1.1023 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1592 -1.8956 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1013 -1.3816 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7898 -1.4124 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5222 -3.2470 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0057 -3.0803 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3890 -1.6096 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8251 -2.5484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6392 -3.3524 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2138 -2.6452 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4874 -2.2536 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6732 -1.4495 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0987 -2.1566 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0355 -1.9820 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2264 -3.3881 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8903 -3.2531 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8059 -3.7703 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3287 -4.1710 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2975 -3.9814 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
2 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 11 1 0 0 0 0
11 12 1 0 0 0 0
12 7 2 0 0 0 0
9 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 10 1 0 0 0 0
14 18 1 0 0 0 0
11 17 2 0 0 0 0
12 19 1 0 0 0 0
16 20 1 0 0 0 0
5 21 1 0 0 0 0
4 22 1 0 0 0 0
22 23 1 0 0 0 0
24 25 1 1 0 0 0
25 26 1 1 0 0 0
27 26 1 1 0 0 0
27 28 1 0 0 0 0
28 29 1 0 0 0 0
29 24 1 0 0 0 0
29 30 1 0 0 0 0
30 31 1 0 0 0 0
24 32 1 0 0 0 0
25 33 1 0 0 0 0
26 34 1 0 0 0 0
27 19 1 0 0 0 0
35 36 1 0 0 0 0
36 37 1 1 0 0 0
38 37 1 1 0 0 0
39 38 1 1 0 0 0
39 40 1 0 0 0 0
40 35 1 0 0 0 0
35 41 1 0 0 0 0
36 42 1 0 0 0 0
37 43 1 0 0 0 0
38 31 1 0 0 0 0
43 44 1 0 0 0 0
44 45 2 0 0 0 0
44 46 1 0 0 0 0
S SKP 5
ID FL5FADGS0030
FORMULA C29H32O17
EXACTMASS 652.163949598
AVERAGEMASS 652.55418
SMILES C(C(O)1)(O)COC(OCC(O2)C(O)C(C(O)C2OC(=C4c(c5)cc(OC)c(O)c5)C(=O)c(c(O4)3)c(cc(O)c3)O)O)C1OC(C)=O
M END