Mol:FL5FADGS0018
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 55 60 0 0 0 0 0 0 0 0999 V2000 | + | 55 60 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.5976 0.5459 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5976 0.5459 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5976 -0.0965 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5976 -0.0965 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0413 -0.4177 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0413 -0.4177 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5150 -0.0965 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5150 -0.0965 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5150 0.5459 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5150 0.5459 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0413 0.8670 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0413 0.8670 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0713 -0.4177 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0713 -0.4177 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6276 -0.0965 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6276 -0.0965 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6276 0.5459 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6276 0.5459 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0713 0.8670 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0713 0.8670 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0713 -0.9185 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0713 -0.9185 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3281 0.9907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3281 0.9907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8951 0.6633 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8951 0.6633 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4621 0.9907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4621 0.9907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4621 1.6454 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4621 1.6454 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8951 1.9727 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8951 1.9727 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3281 1.6454 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3281 1.6454 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0413 -1.0598 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0413 -1.0598 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3281 2.1454 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3281 2.1454 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0925 0.8670 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0925 0.8670 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1909 -0.5060 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1909 -0.5060 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2485 0.7399 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -3.2485 0.7399 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -2.8216 0.1763 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.8216 0.1763 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.2068 0.4154 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.2068 0.4154 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.6135 0.4218 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.6135 0.4218 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.0446 0.8530 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0446 0.8530 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6726 0.6275 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -2.6726 0.6275 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -3.7257 1.0154 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7257 1.0154 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4825 0.2600 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4825 0.2600 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8545 -0.1760 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8545 -0.1760 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1071 -1.2682 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1071 -1.2682 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4534 -0.7800 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -3.4534 -0.7800 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -3.0265 -1.3436 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -3.0265 -1.3436 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.4117 -1.1045 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.4117 -1.1045 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.8185 -1.0981 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.8185 -1.0981 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.2495 -0.6669 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2495 -0.6669 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8775 -0.8924 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -2.8775 -0.8924 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -3.5441 -1.3436 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5441 -1.3436 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0594 -1.6959 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0594 -1.6959 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0580 -1.6204 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 3.0580 -1.6204 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 3.6862 -1.9831 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 3.6862 -1.9831 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.4869 -1.2856 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 3.4869 -1.2856 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 3.6862 -0.5839 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6862 -0.5839 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0580 -0.2211 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 3.0580 -0.2211 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.2573 -0.9187 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 3.2573 -0.9187 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 4.0276 -0.9187 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0276 -0.9187 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2194 -2.2226 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2194 -2.2226 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5347 -2.5488 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5347 -2.5488 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1071 -1.6437 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1071 -1.6437 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9640 -0.4940 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9640 -0.4940 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7609 -0.2805 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7609 -0.2805 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9079 1.3533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9079 1.3533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2383 2.0960 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2383 2.0960 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1785 2.5488 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1785 2.5488 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6199 3.4461 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6199 3.4461 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 20 1 1 0 0 0 0 | + | 20 1 1 0 0 0 0 |
| − | 21 8 1 0 0 0 0 | + | 21 8 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 35 34 1 1 0 0 0 | + | 35 34 1 1 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 33 38 1 0 0 0 0 | + | 33 38 1 0 0 0 0 |
| − | 34 39 1 0 0 0 0 | + | 34 39 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 35 30 1 0 0 0 0 | + | 35 30 1 0 0 0 0 |
| − | 25 20 1 0 0 0 0 | + | 25 20 1 0 0 0 0 |
| − | 41 40 1 1 0 0 0 | + | 41 40 1 1 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 44 45 1 1 0 0 0 | + | 44 45 1 1 0 0 0 |
| − | 45 40 1 1 0 0 0 | + | 45 40 1 1 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | 40 47 1 0 0 0 0 | + | 40 47 1 0 0 0 0 |
| − | 41 48 1 0 0 0 0 | + | 41 48 1 0 0 0 0 |
| − | 42 49 1 0 0 0 0 | + | 42 49 1 0 0 0 0 |
| − | 44 21 1 0 0 0 0 | + | 44 21 1 0 0 0 0 |
| − | 37 50 1 0 0 0 0 | + | 37 50 1 0 0 0 0 |
| − | 50 51 1 0 0 0 0 | + | 50 51 1 0 0 0 0 |
| − | 27 52 1 0 0 0 0 | + | 27 52 1 0 0 0 0 |
| − | 52 53 1 0 0 0 0 | + | 52 53 1 0 0 0 0 |
| − | 16 54 1 0 0 0 0 | + | 16 54 1 0 0 0 0 |
| − | 54 55 1 0 0 0 0 | + | 54 55 1 0 0 0 0 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 52 53 | + | M SAL 3 2 52 53 |
| − | M SBL 3 1 57 | + | M SBL 3 1 57 |
| − | M SMT 3 CH2OH | + | M SMT 3 CH2OH |
| − | M SVB 3 57 -2.9079 1.3533 | + | M SVB 3 57 -2.9079 1.3533 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 50 51 | + | M SAL 2 2 50 51 |
| − | M SBL 2 1 55 | + | M SBL 2 1 55 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SVB 2 55 -2.964 -0.494 | + | M SVB 2 55 -2.964 -0.494 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 54 55 | + | M SAL 1 2 54 55 |
| − | M SBL 1 1 59 | + | M SBL 1 1 59 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 59 3.1785 2.5488 | + | M SVB 1 59 3.1785 2.5488 |
| − | S SKP 8 | + | S SKP 8 |
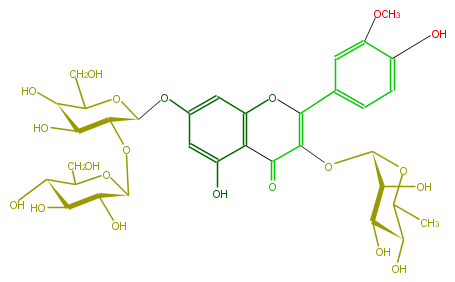
| − | ID FL5FADGS0018 | + | ID FL5FADGS0018 |
| − | KNApSAcK_ID C00005587 | + | KNApSAcK_ID C00005587 |
| − | NAME Isorhamnetin 3-rhamnoside-7-sophoroside | + | NAME Isorhamnetin 3-rhamnoside-7-sophoroside |
| − | CAS_RN 123131-40-8 | + | CAS_RN 123131-40-8 |
| − | FORMULA C34H42O21 | + | FORMULA C34H42O21 |
| − | EXACTMASS 786.2218584059999 | + | EXACTMASS 786.2218584059999 |
| − | AVERAGEMASS 786.68468 | + | AVERAGEMASS 786.68468 |
| − | SMILES O[C@@H]([C@H]6O)C(C)O[C@@H]([C@H]6O)OC(C(=O)1)=C(c(c5)ccc(O)c5OC)Oc(c2)c1c(O)cc2O[C@H](O4)[C@H]([C@H]([C@@H](O)C4CO)O)O[C@H](O3)[C@H]([C@H]([C@H](C3CO)O)O)O | + | SMILES O[C@@H]([C@H]6O)C(C)O[C@@H]([C@H]6O)OC(C(=O)1)=C(c(c5)ccc(O)c5OC)Oc(c2)c1c(O)cc2O[C@H](O4)[C@H]([C@H]([C@@H](O)C4CO)O)O[C@H](O3)[C@H]([C@H]([C@H](C3CO)O)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
55 60 0 0 0 0 0 0 0 0999 V2000
-0.5976 0.5459 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5976 -0.0965 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0413 -0.4177 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5150 -0.0965 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5150 0.5459 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0413 0.8670 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0713 -0.4177 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6276 -0.0965 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6276 0.5459 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0713 0.8670 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0713 -0.9185 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3281 0.9907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8951 0.6633 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4621 0.9907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4621 1.6454 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8951 1.9727 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3281 1.6454 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0413 -1.0598 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.3281 2.1454 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0925 0.8670 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1909 -0.5060 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2485 0.7399 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-2.8216 0.1763 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.2068 0.4154 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.6135 0.4218 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.0446 0.8530 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6726 0.6275 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-3.7257 1.0154 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4825 0.2600 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8545 -0.1760 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1071 -1.2682 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4534 -0.7800 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-3.0265 -1.3436 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.4117 -1.1045 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.8185 -1.0981 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.2495 -0.6669 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8775 -0.8924 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-3.5441 -1.3436 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0594 -1.6959 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0580 -1.6204 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
3.6862 -1.9831 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.4869 -1.2856 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
3.6862 -0.5839 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0580 -0.2211 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.2573 -0.9187 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
4.0276 -0.9187 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2194 -2.2226 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5347 -2.5488 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1071 -1.6437 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9640 -0.4940 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7609 -0.2805 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9079 1.3533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2383 2.0960 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1785 2.5488 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6199 3.4461 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
20 1 1 0 0 0 0
21 8 1 0 0 0 0
4 3 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
24 30 1 0 0 0 0
32 33 1 1 0 0 0
33 34 1 1 0 0 0
35 34 1 1 0 0 0
35 36 1 0 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
33 38 1 0 0 0 0
34 39 1 0 0 0 0
31 32 1 0 0 0 0
35 30 1 0 0 0 0
25 20 1 0 0 0 0
41 40 1 1 0 0 0
41 42 1 0 0 0 0
42 43 1 0 0 0 0
43 44 1 0 0 0 0
44 45 1 1 0 0 0
45 40 1 1 0 0 0
45 46 1 0 0 0 0
40 47 1 0 0 0 0
41 48 1 0 0 0 0
42 49 1 0 0 0 0
44 21 1 0 0 0 0
37 50 1 0 0 0 0
50 51 1 0 0 0 0
27 52 1 0 0 0 0
52 53 1 0 0 0 0
16 54 1 0 0 0 0
54 55 1 0 0 0 0
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 52 53
M SBL 3 1 57
M SMT 3 CH2OH
M SVB 3 57 -2.9079 1.3533
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 50 51
M SBL 2 1 55
M SMT 2 CH2OH
M SVB 2 55 -2.964 -0.494
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 54 55
M SBL 1 1 59
M SMT 1 OCH3
M SVB 1 59 3.1785 2.5488
S SKP 8
ID FL5FADGS0018
KNApSAcK_ID C00005587
NAME Isorhamnetin 3-rhamnoside-7-sophoroside
CAS_RN 123131-40-8
FORMULA C34H42O21
EXACTMASS 786.2218584059999
AVERAGEMASS 786.68468
SMILES O[C@@H]([C@H]6O)C(C)O[C@@H]([C@H]6O)OC(C(=O)1)=C(c(c5)ccc(O)c5OC)Oc(c2)c1c(O)cc2O[C@H](O4)[C@H]([C@H]([C@@H](O)C4CO)O)O[C@H](O3)[C@H]([C@H]([C@H](C3CO)O)O)O
M END