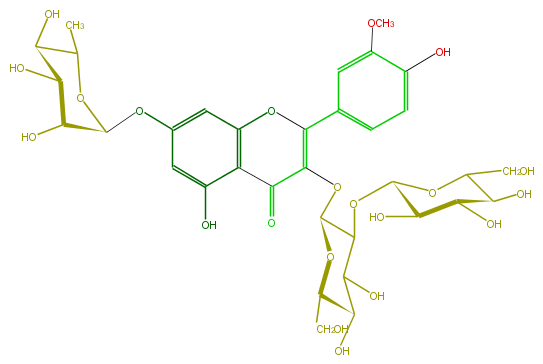
Mol:FL5FADGL0025
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 55 60 0 0 0 0 0 0 0 0999 V2000 | + | 55 60 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.0818 0.6461 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0818 0.6461 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0818 -0.1634 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0818 -0.1634 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3808 -0.5682 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3808 -0.5682 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6798 -0.1634 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6798 -0.1634 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6798 0.6461 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6798 0.6461 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3808 1.0509 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3808 1.0509 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0212 -0.5682 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0212 -0.5682 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7222 -0.1634 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7222 -0.1634 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7222 0.6461 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7222 0.6461 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0212 1.0509 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0212 1.0509 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0226 -1.3244 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0226 -1.3244 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4230 1.0506 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4230 1.0506 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1375 0.6382 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1375 0.6382 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8520 1.0506 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8520 1.0506 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8520 1.8756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8520 1.8756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1375 2.2882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1375 2.2882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4230 1.8756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4230 1.8756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7826 1.0506 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7826 1.0506 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4280 -0.5737 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4280 -0.5737 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3808 -1.3773 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3808 -1.3773 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5734 2.2981 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5734 2.2981 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7731 -1.6494 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7731 -1.6494 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0387 -1.2255 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0387 -1.2255 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2717 -2.0408 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2717 -2.0408 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0387 -2.8612 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0387 -2.8612 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7731 -3.2852 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7731 -3.2852 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5401 -2.4698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5401 -2.4698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7696 -0.9252 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7696 -0.9252 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1880 -2.8850 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1880 -2.8850 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4345 -4.0404 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4345 -4.0404 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5735 -0.4615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5735 -0.4615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1311 -1.1973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1311 -1.1973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9338 -0.8852 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9338 -0.8852 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7084 -0.8769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7084 -0.8769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1455 -0.3138 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1455 -0.3138 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4432 -0.6844 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4432 -0.6844 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3252 -1.1998 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3252 -1.1998 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6633 -1.3494 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6633 -1.3494 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.2988 -0.7148 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.2988 -0.7148 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3921 0.7433 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3921 0.7433 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4919 0.5846 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4919 0.5846 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0287 1.3246 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0287 1.3246 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0950 2.2415 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0950 2.2415 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9952 2.4003 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9952 2.4003 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4587 1.6602 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4587 1.6602 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7054 3.0966 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7054 3.0966 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2804 2.8672 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2804 2.8672 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.2988 1.9510 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.2988 1.9510 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0527 0.4863 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0527 0.4863 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9452 -3.5704 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9452 -3.5704 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3147 -3.5435 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3147 -3.5435 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1454 2.9097 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1454 2.9097 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7016 4.0404 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7016 4.0404 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9893 -0.2217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9893 -0.2217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8215 1.0273 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8215 1.0273 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 8 19 1 0 0 0 0 | + | 8 19 1 0 0 0 0 |
| − | 3 20 1 0 0 0 0 | + | 3 20 1 0 0 0 0 |
| − | 21 15 1 0 0 0 0 | + | 21 15 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 27 29 1 0 0 0 0 | + | 27 29 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 23 19 1 0 0 0 0 | + | 23 19 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 32 37 1 0 0 0 0 | + | 32 37 1 0 0 0 0 |
| − | 33 38 1 0 0 0 0 | + | 33 38 1 0 0 0 0 |
| − | 31 28 1 0 0 0 0 | + | 31 28 1 0 0 0 0 |
| − | 34 39 1 0 0 0 0 | + | 34 39 1 0 0 0 0 |
| − | 41 40 1 1 0 0 0 | + | 41 40 1 1 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 44 45 1 1 0 0 0 | + | 44 45 1 1 0 0 0 |
| − | 45 40 1 1 0 0 0 | + | 45 40 1 1 0 0 0 |
| − | 44 46 1 0 0 0 0 | + | 44 46 1 0 0 0 0 |
| − | 43 47 1 0 0 0 0 | + | 43 47 1 0 0 0 0 |
| − | 45 48 1 0 0 0 0 | + | 45 48 1 0 0 0 0 |
| − | 40 49 1 0 0 0 0 | + | 40 49 1 0 0 0 0 |
| − | 41 18 1 0 0 0 0 | + | 41 18 1 0 0 0 0 |
| − | 50 51 1 0 0 0 0 | + | 50 51 1 0 0 0 0 |
| − | 25 50 1 0 0 0 0 | + | 25 50 1 0 0 0 0 |
| − | 52 53 1 0 0 0 0 | + | 52 53 1 0 0 0 0 |
| − | 16 52 1 0 0 0 0 | + | 16 52 1 0 0 0 0 |
| − | 54 55 1 0 0 0 0 | + | 54 55 1 0 0 0 0 |
| − | 35 54 1 0 0 0 0 | + | 35 54 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 50 51 | + | M SAL 1 2 50 51 |
| − | M SBL 1 1 56 | + | M SBL 1 1 56 |
| − | M SMT 1 ^ CH2OH | + | M SMT 1 ^ CH2OH |
| − | M SBV 1 56 0.0935 0.7092 | + | M SBV 1 56 0.0935 0.7092 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 52 53 | + | M SAL 2 2 52 53 |
| − | M SBL 2 1 58 | + | M SBL 2 1 58 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SBV 2 58 -0.0079 -0.6215 | + | M SBV 2 58 -0.0079 -0.6215 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 54 55 | + | M SAL 3 2 54 55 |
| − | M SBL 3 1 60 | + | M SBL 3 1 60 |
| − | M SMT 3 CH2OH | + | M SMT 3 CH2OH |
| − | M SBV 3 60 -0.8438 -0.0922 | + | M SBV 3 60 -0.8438 -0.0922 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FADGL0025 | + | ID FL5FADGL0025 |
| − | FORMULA C34H42O21 | + | FORMULA C34H42O21 |
| − | EXACTMASS 786.2218584059999 | + | EXACTMASS 786.2218584059999 |
| − | AVERAGEMASS 786.68468 | + | AVERAGEMASS 786.68468 |
| − | SMILES C(C1O)(C(OC(C2O)C(OC(=C(c(c6)ccc(c6OC)O)5)C(c(c4O5)c(cc(c4)OC(C3O)OC(C(O)C3O)C)O)=O)OC(C2O)CO)OC(C1O)CO)O | + | SMILES C(C1O)(C(OC(C2O)C(OC(=C(c(c6)ccc(c6OC)O)5)C(c(c4O5)c(cc(c4)OC(C3O)OC(C(O)C3O)C)O)=O)OC(C2O)CO)OC(C1O)CO)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
55 60 0 0 0 0 0 0 0 0999 V2000
-2.0818 0.6461 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0818 -0.1634 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3808 -0.5682 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6798 -0.1634 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6798 0.6461 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3808 1.0509 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0212 -0.5682 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7222 -0.1634 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7222 0.6461 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0212 1.0509 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0226 -1.3244 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4230 1.0506 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1375 0.6382 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8520 1.0506 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8520 1.8756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1375 2.2882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4230 1.8756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7826 1.0506 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4280 -0.5737 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3808 -1.3773 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5734 2.2981 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7731 -1.6494 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0387 -1.2255 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2717 -2.0408 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0387 -2.8612 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7731 -3.2852 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5401 -2.4698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7696 -0.9252 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1880 -2.8850 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4345 -4.0404 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5735 -0.4615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1311 -1.1973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9338 -0.8852 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.7084 -0.8769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1455 -0.3138 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4432 -0.6844 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3252 -1.1998 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.6633 -1.3494 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.2988 -0.7148 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.3921 0.7433 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4919 0.5846 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0287 1.3246 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0950 2.2415 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.9952 2.4003 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4587 1.6602 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.7054 3.0966 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2804 2.8672 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.2988 1.9510 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.0527 0.4863 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9452 -3.5704 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3147 -3.5435 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1454 2.9097 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7016 4.0404 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.9893 -0.2217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.8215 1.0273 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
8 19 1 0 0 0 0
3 20 1 0 0 0 0
21 15 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
27 29 1 0 0 0 0
26 30 1 0 0 0 0
23 19 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
32 37 1 0 0 0 0
33 38 1 0 0 0 0
31 28 1 0 0 0 0
34 39 1 0 0 0 0
41 40 1 1 0 0 0
41 42 1 0 0 0 0
42 43 1 0 0 0 0
43 44 1 0 0 0 0
44 45 1 1 0 0 0
45 40 1 1 0 0 0
44 46 1 0 0 0 0
43 47 1 0 0 0 0
45 48 1 0 0 0 0
40 49 1 0 0 0 0
41 18 1 0 0 0 0
50 51 1 0 0 0 0
25 50 1 0 0 0 0
52 53 1 0 0 0 0
16 52 1 0 0 0 0
54 55 1 0 0 0 0
35 54 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 50 51
M SBL 1 1 56
M SMT 1 ^ CH2OH
M SBV 1 56 0.0935 0.7092
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 52 53
M SBL 2 1 58
M SMT 2 OCH3
M SBV 2 58 -0.0079 -0.6215
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 54 55
M SBL 3 1 60
M SMT 3 CH2OH
M SBV 3 60 -0.8438 -0.0922
S SKP 5
ID FL5FADGL0025
FORMULA C34H42O21
EXACTMASS 786.2218584059999
AVERAGEMASS 786.68468
SMILES C(C1O)(C(OC(C2O)C(OC(=C(c(c6)ccc(c6OC)O)5)C(c(c4O5)c(cc(c4)OC(C3O)OC(C(O)C3O)C)O)=O)OC(C2O)CO)OC(C1O)CO)O
M END