Mol:FL5FACGL0078
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 55 60 0 0 0 0 0 0 0 0999 V2000 | + | 55 60 0 0 0 0 0 0 0 0999 V2000 |
| − | 0.0739 1.1629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0739 1.1629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7007 1.5248 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7007 1.5248 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7007 2.2486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7007 2.2486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0739 2.6104 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0739 2.6104 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5528 2.2486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5528 2.2486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5528 1.5248 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5528 1.5248 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3275 1.1629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3275 1.1629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9543 1.5248 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9543 1.5248 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9543 2.2486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9543 2.2486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3275 2.6104 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3275 2.6104 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5837 2.6119 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5837 2.6119 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1947 2.2591 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1947 2.2591 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8058 2.6119 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8058 2.6119 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8058 3.3175 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8058 3.3175 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1947 3.6703 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1947 3.6703 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5837 3.3175 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5837 3.3175 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3275 0.5687 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3275 0.5687 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2479 3.5728 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2479 3.5728 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1947 4.1834 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1947 4.1834 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0739 0.6339 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0739 0.6339 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4346 1.2475 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4346 1.2475 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0741 2.5495 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0741 2.5495 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7978 2.6450 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -3.7978 2.6450 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -3.2184 2.0577 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -3.2184 2.0577 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.4989 2.4614 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.4989 2.4614 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.6741 2.4421 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.6741 2.4421 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.2535 3.0295 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2535 3.0295 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9730 2.6258 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -2.9730 2.6258 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -3.2561 3.1161 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2561 3.1161 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9378 3.4344 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9378 3.4344 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2479 2.1948 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2479 2.1948 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7131 2.1903 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7131 2.1903 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3717 1.9869 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3717 1.9869 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1669 -0.8311 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 1.1669 -0.8311 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 1.9887 -0.9042 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.9887 -0.9042 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.2763 -0.1309 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.2763 -0.1309 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.9178 0.3878 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.9178 0.3878 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.0960 0.4610 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0960 0.4610 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8083 -0.3123 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 1.8083 -0.3123 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 2.7796 -0.4216 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7796 -0.4216 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1669 -1.4774 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1669 -1.4774 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5426 -1.1618 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5426 -1.1618 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2611 0.0037 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2611 0.0037 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7318 -0.3019 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7318 -0.3019 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1993 -0.8344 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1993 -0.8344 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3532 -0.2819 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3532 -0.2819 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1993 -1.4276 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1993 -1.4276 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4219 -1.7862 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4219 -1.7862 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4219 -2.3518 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4219 -2.3518 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0030 -2.6873 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0030 -2.6873 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0030 -3.3583 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0030 -3.3583 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4219 -3.6938 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4219 -3.6938 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1592 -3.3583 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1592 -3.3583 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1592 -2.6873 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1592 -2.6873 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4219 -4.1834 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4219 -4.1834 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 2 7 1 0 0 0 0 | + | 2 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 3 1 0 0 0 0 | + | 10 3 1 0 0 0 0 |
| − | 9 11 1 0 0 0 0 | + | 9 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 7 17 2 0 0 0 0 | + | 7 17 2 0 0 0 0 |
| − | 14 18 1 0 0 0 0 | + | 14 18 1 0 0 0 0 |
| − | 15 19 1 0 0 0 0 | + | 15 19 1 0 0 0 0 |
| − | 1 20 1 0 0 0 0 | + | 1 20 1 0 0 0 0 |
| − | 8 21 1 0 0 0 0 | + | 8 21 1 0 0 0 0 |
| − | 5 22 1 0 0 0 0 | + | 5 22 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 23 1 0 0 0 0 | + | 28 23 1 0 0 0 0 |
| − | 22 26 1 0 0 0 0 | + | 22 26 1 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 29 30 1 0 0 0 0 | + | 29 30 1 0 0 0 0 |
| − | 23 31 1 0 0 0 0 | + | 23 31 1 0 0 0 0 |
| − | 24 32 1 0 0 0 0 | + | 24 32 1 0 0 0 0 |
| − | 25 33 1 0 0 0 0 | + | 25 33 1 0 0 0 0 |
| − | 34 35 1 1 0 0 0 | + | 34 35 1 1 0 0 0 |
| − | 35 36 1 1 0 0 0 | + | 35 36 1 1 0 0 0 |
| − | 37 36 1 1 0 0 0 | + | 37 36 1 1 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 39 34 1 0 0 0 0 | + | 39 34 1 0 0 0 0 |
| − | 21 37 1 0 0 0 0 | + | 21 37 1 0 0 0 0 |
| − | 36 40 1 0 0 0 0 | + | 36 40 1 0 0 0 0 |
| − | 34 41 1 0 0 0 0 | + | 34 41 1 0 0 0 0 |
| − | 35 42 1 0 0 0 0 | + | 35 42 1 0 0 0 0 |
| − | 39 43 1 0 0 0 0 | + | 39 43 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 45 46 2 0 0 0 0 | + | 45 46 2 0 0 0 0 |
| − | 45 47 1 0 0 0 0 | + | 45 47 1 0 0 0 0 |
| − | 47 48 2 0 0 0 0 | + | 47 48 2 0 0 0 0 |
| − | 52 55 1 0 0 0 0 | + | 52 55 1 0 0 0 0 |
| − | 54 49 1 0 0 0 0 | + | 54 49 1 0 0 0 0 |
| − | 48 49 1 0 0 0 0 | + | 48 49 1 0 0 0 0 |
| − | 49 50 2 0 0 0 0 | + | 49 50 2 0 0 0 0 |
| − | 50 51 1 0 0 0 0 | + | 50 51 1 0 0 0 0 |
| − | 51 52 2 0 0 0 0 | + | 51 52 2 0 0 0 0 |
| − | 52 53 1 0 0 0 0 | + | 52 53 1 0 0 0 0 |
| − | 53 54 2 0 0 0 0 | + | 53 54 2 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 29 30 | + | M SAL 1 2 29 30 |
| − | M SBL 1 1 32 | + | M SBL 1 1 32 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SVB 1 32 -3.2561 3.1161 | + | M SVB 1 32 -3.2561 3.1161 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FACGL0078 | + | ID FL5FACGL0078 |
| − | KNApSAcK_ID C00011129 | + | KNApSAcK_ID C00011129 |
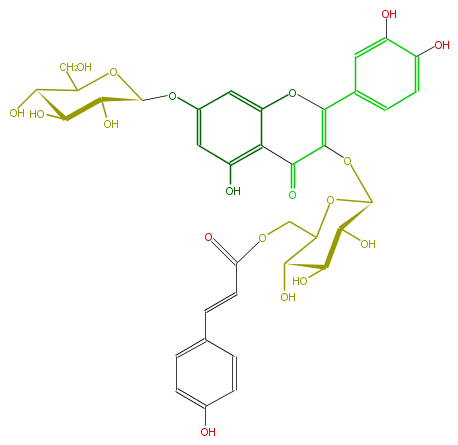
| − | NAME Quercetin 3-O-beta-(6''-O-E-p-coumaroylglucoside)-7-O-beta-glucoside | + | NAME Quercetin 3-O-beta-(6''-O-E-p-coumaroylglucoside)-7-O-beta-glucoside |
| − | CAS_RN 473919-25-4 | + | CAS_RN 473919-25-4 |
| − | FORMULA C36H36O19 | + | FORMULA C36H36O19 |
| − | EXACTMASS 772.18507897 | + | EXACTMASS 772.18507897 |
| − | AVERAGEMASS 772.6596400000001 | + | AVERAGEMASS 772.6596400000001 |
| − | SMILES C(=O)(OCC([C@@H]6O)O[C@H]([C@H]([C@@H](O)6)O)OC(=C(c(c5)ccc(O)c5O)4)C(c(c(O4)2)c(O)cc(O[C@H](O3)[C@@H](O)[C@H]([C@@H](O)C(CO)3)O)c2)=O)C=Cc(c1)ccc(c1)O | + | SMILES C(=O)(OCC([C@@H]6O)O[C@H]([C@H]([C@@H](O)6)O)OC(=C(c(c5)ccc(O)c5O)4)C(c(c(O4)2)c(O)cc(O[C@H](O3)[C@@H](O)[C@H]([C@@H](O)C(CO)3)O)c2)=O)C=Cc(c1)ccc(c1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
55 60 0 0 0 0 0 0 0 0999 V2000
0.0739 1.1629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7007 1.5248 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7007 2.2486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0739 2.6104 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5528 2.2486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5528 1.5248 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3275 1.1629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9543 1.5248 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9543 2.2486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3275 2.6104 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5837 2.6119 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1947 2.2591 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8058 2.6119 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8058 3.3175 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1947 3.6703 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5837 3.3175 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3275 0.5687 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.2479 3.5728 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1947 4.1834 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0739 0.6339 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4346 1.2475 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0741 2.5495 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7978 2.6450 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-3.2184 2.0577 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.4989 2.4614 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.6741 2.4421 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.2535 3.0295 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9730 2.6258 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-3.2561 3.1161 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9378 3.4344 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2479 2.1948 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7131 2.1903 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3717 1.9869 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1669 -0.8311 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
1.9887 -0.9042 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.2763 -0.1309 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.9178 0.3878 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.0960 0.4610 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8083 -0.3123 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
2.7796 -0.4216 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1669 -1.4774 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5426 -1.1618 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2611 0.0037 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7318 -0.3019 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1993 -0.8344 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3532 -0.2819 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1993 -1.4276 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4219 -1.7862 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4219 -2.3518 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0030 -2.6873 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0030 -3.3583 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4219 -3.6938 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1592 -3.3583 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1592 -2.6873 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4219 -4.1834 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
2 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 3 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
7 17 2 0 0 0 0
14 18 1 0 0 0 0
15 19 1 0 0 0 0
1 20 1 0 0 0 0
8 21 1 0 0 0 0
5 22 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 28 1 0 0 0 0
28 23 1 0 0 0 0
22 26 1 0 0 0 0
28 29 1 0 0 0 0
29 30 1 0 0 0 0
23 31 1 0 0 0 0
24 32 1 0 0 0 0
25 33 1 0 0 0 0
34 35 1 1 0 0 0
35 36 1 1 0 0 0
37 36 1 1 0 0 0
37 38 1 0 0 0 0
38 39 1 0 0 0 0
39 34 1 0 0 0 0
21 37 1 0 0 0 0
36 40 1 0 0 0 0
34 41 1 0 0 0 0
35 42 1 0 0 0 0
39 43 1 0 0 0 0
43 44 1 0 0 0 0
44 45 1 0 0 0 0
45 46 2 0 0 0 0
45 47 1 0 0 0 0
47 48 2 0 0 0 0
52 55 1 0 0 0 0
54 49 1 0 0 0 0
48 49 1 0 0 0 0
49 50 2 0 0 0 0
50 51 1 0 0 0 0
51 52 2 0 0 0 0
52 53 1 0 0 0 0
53 54 2 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 29 30
M SBL 1 1 32
M SMT 1 CH2OH
M SVB 1 32 -3.2561 3.1161
S SKP 8
ID FL5FACGL0078
KNApSAcK_ID C00011129
NAME Quercetin 3-O-beta-(6''-O-E-p-coumaroylglucoside)-7-O-beta-glucoside
CAS_RN 473919-25-4
FORMULA C36H36O19
EXACTMASS 772.18507897
AVERAGEMASS 772.6596400000001
SMILES C(=O)(OCC([C@@H]6O)O[C@H]([C@H]([C@@H](O)6)O)OC(=C(c(c5)ccc(O)c5O)4)C(c(c(O4)2)c(O)cc(O[C@H](O3)[C@@H](O)[C@H]([C@@H](O)C(CO)3)O)c2)=O)C=Cc(c1)ccc(c1)O
M END