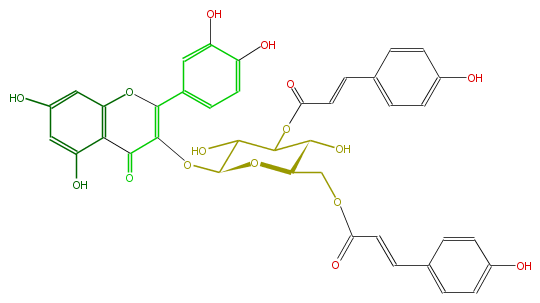
Mol:FL5FACGL0061
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 55 60 0 0 0 0 0 0 0 0999 V2000 | + | 55 60 0 0 0 0 0 0 0 0999 V2000 |
| − | -4.6005 0.9555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6005 0.9555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6005 0.3131 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6005 0.3131 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0442 -0.0081 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0442 -0.0081 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4879 0.3131 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4879 0.3131 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4879 0.9555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4879 0.9555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0442 1.2767 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0442 1.2767 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9316 -0.0081 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9316 -0.0081 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3753 0.3131 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3753 0.3131 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3753 0.9555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3753 0.9555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9316 1.2767 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9316 1.2767 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9316 -0.5089 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9316 -0.5089 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8192 1.2765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8192 1.2765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2522 0.9492 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2522 0.9492 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6852 1.2765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6852 1.2765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6852 1.9312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6852 1.9312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2522 2.2586 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2522 2.2586 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8192 1.9312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8192 1.9312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7272 -0.2371 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7272 -0.2371 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6817 0.2542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6817 0.2542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0694 -0.4172 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0694 -0.4172 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3239 -0.2041 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3239 -0.2041 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4261 -0.4172 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4261 -0.4172 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8138 0.2542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8138 0.2542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0683 0.0413 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0683 0.0413 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4088 0.0594 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4088 0.0594 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3371 0.5069 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3371 0.5069 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4281 0.0896 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4281 0.0896 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1184 2.2585 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1184 2.2585 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0442 -0.6502 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0442 -0.6502 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.2114 1.1540 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.2114 1.1540 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0550 -0.4172 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0550 -0.4172 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4006 -1.0157 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4006 -1.0157 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6902 -1.7464 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6902 -1.7464 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3576 -2.3224 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3576 -2.3224 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2522 2.9131 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2522 2.9131 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2268 -1.7464 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2268 -1.7464 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5787 -2.3559 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5787 -2.3559 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2825 -2.3559 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2825 -2.3559 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6042 -2.9131 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6042 -2.9131 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2476 -2.9131 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2476 -2.9131 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5693 -2.3559 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5693 -2.3559 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2476 -1.7987 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2476 -1.7987 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6042 -1.7987 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6042 -1.7987 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.2114 -2.3559 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.2114 -2.3559 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6420 1.0349 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6420 1.0349 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4079 1.4403 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4079 1.4403 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2511 1.0349 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2511 1.0349 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5557 1.5624 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5557 1.5624 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1634 1.5624 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1634 1.5624 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4997 0.9799 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4997 0.9799 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1723 0.9799 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1723 0.9799 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5086 1.5624 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5086 1.5624 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1723 2.1449 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1723 2.1449 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4997 2.1449 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4997 2.1449 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1810 1.5624 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1810 1.5624 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 19 20 1 0 0 0 0 | + | 19 20 1 0 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 19 1 0 0 0 0 | + | 24 19 1 0 0 0 0 |
| − | 19 25 1 0 0 0 0 | + | 19 25 1 0 0 0 0 |
| − | 24 26 1 0 0 0 0 | + | 24 26 1 0 0 0 0 |
| − | 23 27 1 0 0 0 0 | + | 23 27 1 0 0 0 0 |
| − | 20 18 1 0 0 0 0 | + | 20 18 1 0 0 0 0 |
| − | 18 8 1 0 0 0 0 | + | 18 8 1 0 0 0 0 |
| − | 22 21 1 1 0 0 0 | + | 22 21 1 1 0 0 0 |
| − | 15 28 1 0 0 0 0 | + | 15 28 1 0 0 0 0 |
| − | 3 29 1 0 0 0 0 | + | 3 29 1 0 0 0 0 |
| − | 1 30 1 0 0 0 0 | + | 1 30 1 0 0 0 0 |
| − | 22 31 1 0 0 0 0 | + | 22 31 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 2 0 0 0 0 | + | 33 34 2 0 0 0 0 |
| − | 16 35 1 0 0 0 0 | + | 16 35 1 0 0 0 0 |
| − | 33 36 1 0 0 0 0 | + | 33 36 1 0 0 0 0 |
| − | 36 37 2 0 0 0 0 | + | 36 37 2 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 2 0 0 0 0 | + | 38 39 2 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 40 41 2 0 0 0 0 | + | 40 41 2 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 42 43 2 0 0 0 0 | + | 42 43 2 0 0 0 0 |
| − | 43 38 1 0 0 0 0 | + | 43 38 1 0 0 0 0 |
| − | 41 44 1 0 0 0 0 | + | 41 44 1 0 0 0 0 |
| − | 26 45 1 0 0 0 0 | + | 26 45 1 0 0 0 0 |
| − | 45 46 2 0 0 0 0 | + | 45 46 2 0 0 0 0 |
| − | 45 47 1 0 0 0 0 | + | 45 47 1 0 0 0 0 |
| − | 47 48 2 0 0 0 0 | + | 47 48 2 0 0 0 0 |
| − | 48 49 1 0 0 0 0 | + | 48 49 1 0 0 0 0 |
| − | 49 50 2 0 0 0 0 | + | 49 50 2 0 0 0 0 |
| − | 50 51 1 0 0 0 0 | + | 50 51 1 0 0 0 0 |
| − | 51 52 2 0 0 0 0 | + | 51 52 2 0 0 0 0 |
| − | 52 53 1 0 0 0 0 | + | 52 53 1 0 0 0 0 |
| − | 53 54 2 0 0 0 0 | + | 53 54 2 0 0 0 0 |
| − | 54 49 1 0 0 0 0 | + | 54 49 1 0 0 0 0 |
| − | 52 55 1 0 0 0 0 | + | 52 55 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FACGL0061 | + | ID FL5FACGL0061 |
| − | KNApSAcK_ID C00005963 | + | KNApSAcK_ID C00005963 |
| − | NAME Quercetin 3-(3'',6''-di-p-coumarylglucoside) | + | NAME Quercetin 3-(3'',6''-di-p-coumarylglucoside) |
| − | CAS_RN 72691-78-2 | + | CAS_RN 72691-78-2 |
| − | FORMULA C39H32O16 | + | FORMULA C39H32O16 |
| − | EXACTMASS 756.1690349759999 | + | EXACTMASS 756.1690349759999 |
| − | AVERAGEMASS 756.66178 | + | AVERAGEMASS 756.66178 |
| − | SMILES O(C(C2O)C(C(OC(C(=O)6)=C(Oc(c65)cc(cc5O)O)c(c4)ccc(c4O)O)OC2COC(C=Cc(c3)ccc(c3)O)=O)O)C(=O)C=Cc(c1)ccc(c1)O | + | SMILES O(C(C2O)C(C(OC(C(=O)6)=C(Oc(c65)cc(cc5O)O)c(c4)ccc(c4O)O)OC2COC(C=Cc(c3)ccc(c3)O)=O)O)C(=O)C=Cc(c1)ccc(c1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
55 60 0 0 0 0 0 0 0 0999 V2000
-4.6005 0.9555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.6005 0.3131 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0442 -0.0081 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4879 0.3131 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4879 0.9555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0442 1.2767 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9316 -0.0081 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3753 0.3131 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3753 0.9555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9316 1.2767 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9316 -0.5089 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8192 1.2765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2522 0.9492 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6852 1.2765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6852 1.9312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2522 2.2586 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8192 1.9312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7272 -0.2371 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6817 0.2542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0694 -0.4172 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3239 -0.2041 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4261 -0.4172 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8138 0.2542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0683 0.0413 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4088 0.0594 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3371 0.5069 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4281 0.0896 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1184 2.2585 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0442 -0.6502 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.2114 1.1540 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0550 -0.4172 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4006 -1.0157 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6902 -1.7464 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3576 -2.3224 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2522 2.9131 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2268 -1.7464 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5787 -2.3559 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2825 -2.3559 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6042 -2.9131 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2476 -2.9131 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5693 -2.3559 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2476 -1.7987 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6042 -1.7987 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.2114 -2.3559 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6420 1.0349 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4079 1.4403 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2511 1.0349 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5557 1.5624 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1634 1.5624 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4997 0.9799 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1723 0.9799 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5086 1.5624 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1723 2.1449 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4997 2.1449 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1810 1.5624 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
19 20 1 0 0 0 0
20 21 1 1 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 19 1 0 0 0 0
19 25 1 0 0 0 0
24 26 1 0 0 0 0
23 27 1 0 0 0 0
20 18 1 0 0 0 0
18 8 1 0 0 0 0
22 21 1 1 0 0 0
15 28 1 0 0 0 0
3 29 1 0 0 0 0
1 30 1 0 0 0 0
22 31 1 0 0 0 0
31 32 1 0 0 0 0
32 33 1 0 0 0 0
33 34 2 0 0 0 0
16 35 1 0 0 0 0
33 36 1 0 0 0 0
36 37 2 0 0 0 0
37 38 1 0 0 0 0
38 39 2 0 0 0 0
39 40 1 0 0 0 0
40 41 2 0 0 0 0
41 42 1 0 0 0 0
42 43 2 0 0 0 0
43 38 1 0 0 0 0
41 44 1 0 0 0 0
26 45 1 0 0 0 0
45 46 2 0 0 0 0
45 47 1 0 0 0 0
47 48 2 0 0 0 0
48 49 1 0 0 0 0
49 50 2 0 0 0 0
50 51 1 0 0 0 0
51 52 2 0 0 0 0
52 53 1 0 0 0 0
53 54 2 0 0 0 0
54 49 1 0 0 0 0
52 55 1 0 0 0 0
S SKP 8
ID FL5FACGL0061
KNApSAcK_ID C00005963
NAME Quercetin 3-(3'',6''-di-p-coumarylglucoside)
CAS_RN 72691-78-2
FORMULA C39H32O16
EXACTMASS 756.1690349759999
AVERAGEMASS 756.66178
SMILES O(C(C2O)C(C(OC(C(=O)6)=C(Oc(c65)cc(cc5O)O)c(c4)ccc(c4O)O)OC2COC(C=Cc(c3)ccc(c3)O)=O)O)C(=O)C=Cc(c1)ccc(c1)O
M END