Mol:FL5FABGI0016
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 38 41 0 0 0 0 0 0 0 0999 V2000 | + | 38 41 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.4246 0.2789 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4246 0.2789 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4246 -0.3635 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4246 -0.3635 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8683 -0.6847 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8683 -0.6847 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3120 -0.3635 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3120 -0.3635 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3120 0.2789 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3120 0.2789 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8683 0.6000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8683 0.6000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7557 -0.6847 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7557 -0.6847 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1994 -0.3635 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1994 -0.3635 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1994 0.2789 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1994 0.2789 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7557 0.6000 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7557 0.6000 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7557 -1.1855 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7557 -1.1855 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3567 0.5999 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3567 0.5999 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9237 0.2726 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9237 0.2726 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4906 0.5999 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4906 0.5999 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4906 1.2546 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4906 1.2546 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9237 1.5819 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9237 1.5819 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3567 1.2546 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3567 1.2546 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8683 -1.3268 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8683 -1.3268 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3567 -0.6846 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3567 -0.6846 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4166 -1.8049 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4166 -1.8049 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0448 -2.1676 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0448 -2.1676 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8455 -1.4701 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8455 -1.4701 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0448 -0.7684 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0448 -0.7684 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4166 -0.4056 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4166 -0.4056 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6159 -1.1032 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6159 -1.1032 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3862 -1.1032 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3862 -1.1032 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5780 -2.4072 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5780 -2.4072 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8932 -2.7333 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8932 -2.7333 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4657 -1.8282 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4657 -1.8282 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8683 1.2101 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8683 1.2101 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3966 1.5151 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3966 1.5151 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3966 2.1242 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3966 2.1242 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9241 2.4288 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9241 2.4288 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8691 2.4288 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8691 2.4288 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3966 2.7333 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3966 2.7333 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9807 0.5999 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9807 0.5999 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2662 1.6345 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2662 1.6345 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9807 1.2220 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9807 1.2220 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 8 19 1 0 0 0 0 | + | 8 19 1 0 0 0 0 |
| − | 21 20 1 1 0 0 0 | + | 21 20 1 1 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 25 20 1 1 0 0 0 | + | 25 20 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 20 27 1 0 0 0 0 | + | 20 27 1 0 0 0 0 |
| − | 21 28 1 0 0 0 0 | + | 21 28 1 0 0 0 0 |
| − | 22 29 1 0 0 0 0 | + | 22 29 1 0 0 0 0 |
| − | 24 19 1 0 0 0 0 | + | 24 19 1 0 0 0 0 |
| − | 6 30 1 0 0 0 0 | + | 6 30 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 32 34 1 0 0 0 0 | + | 32 34 1 0 0 0 0 |
| − | 32 35 1 0 0 0 0 | + | 32 35 1 0 0 0 0 |
| − | 1 36 1 0 0 0 0 | + | 1 36 1 0 0 0 0 |
| − | 15 37 1 0 0 0 0 | + | 15 37 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 37 38 | + | M SAL 1 2 37 38 |
| − | M SBL 1 1 40 | + | M SBL 1 1 40 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 40 -5.7286 4.9075 | + | M SBV 1 40 -5.7286 4.9075 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FABGI0016 | + | ID FL5FABGI0016 |
| − | KNApSAcK_ID C00005833 | + | KNApSAcK_ID C00005833 |
| − | NAME Icaritin 3-rhamnoside | + | NAME Icaritin 3-rhamnoside |
| − | CAS_RN 108195-76-2 | + | CAS_RN 108195-76-2 |
| − | FORMULA C27H32O11 | + | FORMULA C27H32O11 |
| − | EXACTMASS 532.194461866 | + | EXACTMASS 532.194461866 |
| − | AVERAGEMASS 532.53638 | + | AVERAGEMASS 532.53638 |
| − | SMILES c(c(O)1)c(O)c(c(O3)c1C(C(=C3c(c4)ccc(OC)c4)OC(C(O)2)OC(C)C(C(O)2)O)=O)CCC(C)(C)O | + | SMILES c(c(O)1)c(O)c(c(O3)c1C(C(=C3c(c4)ccc(OC)c4)OC(C(O)2)OC(C)C(C(O)2)O)=O)CCC(C)(C)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
38 41 0 0 0 0 0 0 0 0999 V2000
-2.4246 0.2789 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4246 -0.3635 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8683 -0.6847 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3120 -0.3635 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3120 0.2789 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8683 0.6000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7557 -0.6847 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1994 -0.3635 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1994 0.2789 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7557 0.6000 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7557 -1.1855 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3567 0.5999 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9237 0.2726 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4906 0.5999 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4906 1.2546 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9237 1.5819 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3567 1.2546 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8683 -1.3268 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3567 -0.6846 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4166 -1.8049 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0448 -2.1676 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8455 -1.4701 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0448 -0.7684 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4166 -0.4056 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6159 -1.1032 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3862 -1.1032 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5780 -2.4072 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8932 -2.7333 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4657 -1.8282 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8683 1.2101 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3966 1.5151 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3966 2.1242 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9241 2.4288 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8691 2.4288 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3966 2.7333 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9807 0.5999 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2662 1.6345 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9807 1.2220 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
8 19 1 0 0 0 0
21 20 1 1 0 0 0
21 22 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 0 0 0 0
24 25 1 1 0 0 0
25 20 1 1 0 0 0
25 26 1 0 0 0 0
20 27 1 0 0 0 0
21 28 1 0 0 0 0
22 29 1 0 0 0 0
24 19 1 0 0 0 0
6 30 1 0 0 0 0
30 31 1 0 0 0 0
31 32 1 0 0 0 0
32 33 1 0 0 0 0
32 34 1 0 0 0 0
32 35 1 0 0 0 0
1 36 1 0 0 0 0
15 37 1 0 0 0 0
37 38 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 37 38
M SBL 1 1 40
M SMT 1 OCH3
M SBV 1 40 -5.7286 4.9075
S SKP 8
ID FL5FABGI0016
KNApSAcK_ID C00005833
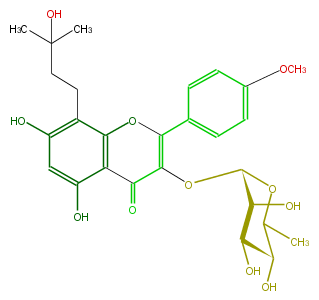
NAME Icaritin 3-rhamnoside
CAS_RN 108195-76-2
FORMULA C27H32O11
EXACTMASS 532.194461866
AVERAGEMASS 532.53638
SMILES c(c(O)1)c(O)c(c(O3)c1C(C(=C3c(c4)ccc(OC)c4)OC(C(O)2)OC(C)C(C(O)2)O)=O)CCC(C)(C)O
M END