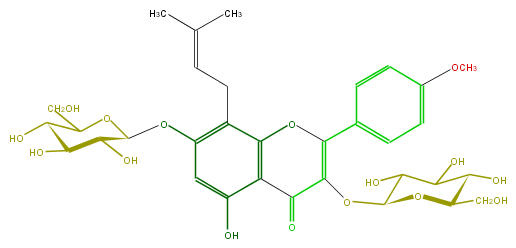
Mol:FL5FABGI0006
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 49 53 0 0 0 0 0 0 0 0999 V2000 | + | 49 53 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.6518 -0.4123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6518 -0.4123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6518 -1.2377 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6518 -1.2377 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9370 -1.6504 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9370 -1.6504 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2223 -1.2377 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2223 -1.2377 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2223 -0.4123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2223 -0.4123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9370 0.0003 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9370 0.0003 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4925 -1.6504 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4925 -1.6504 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2073 -1.2377 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2073 -1.2377 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2073 -0.4123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2073 -0.4123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4925 0.0003 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4925 0.0003 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4925 -2.2938 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4925 -2.2938 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9218 0.0001 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9218 0.0001 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6503 -0.4204 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6503 -0.4204 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3788 0.0001 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3788 0.0001 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3788 0.8413 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3788 0.8413 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6503 1.2618 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6503 1.2618 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9218 0.8413 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9218 0.8413 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3663 0.0001 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3663 0.0001 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7137 -1.7374 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7137 -1.7374 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9370 -2.4754 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9370 -2.4754 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9370 0.8254 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9370 0.8254 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6515 1.2378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6515 1.2378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6515 2.0628 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6515 2.0628 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9370 2.4754 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9370 2.4754 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3660 2.4754 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3660 2.4754 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9680 0.0141 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9680 0.0141 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4910 -0.6155 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4910 -0.6155 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8043 -0.3484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8043 -0.3484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1417 -0.3413 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1417 -0.3413 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6231 0.1404 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6231 0.1404 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2239 -0.1767 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2239 -0.1767 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.5649 -0.3066 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.5649 -0.3066 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.1093 -0.6325 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.1093 -0.6325 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1825 -0.8040 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1825 -0.8040 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9431 -1.1421 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9431 -1.1421 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5566 -1.8116 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5566 -1.8116 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2999 -1.5991 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2999 -1.5991 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0477 -1.8116 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0477 -1.8116 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4343 -1.1421 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4343 -1.1421 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6909 -1.3545 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6909 -1.3545 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3504 -1.2946 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3504 -1.2946 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1014 -0.8259 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1014 -0.8259 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0345 -1.2592 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0345 -1.2592 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1070 1.2618 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1070 1.2618 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0249 0.7318 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0249 0.7318 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6468 -1.6962 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6468 -1.6962 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.5649 -2.2262 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.5649 -2.2262 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8775 0.3537 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8775 0.3537 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8536 0.0794 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8536 0.0794 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 8 19 1 0 0 0 0 | + | 8 19 1 0 0 0 0 |
| − | 3 20 1 0 0 0 0 | + | 3 20 1 0 0 0 0 |
| − | 6 21 1 0 0 0 0 | + | 6 21 1 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 22 23 2 0 0 0 0 | + | 22 23 2 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 23 25 1 0 0 0 0 | + | 23 25 1 0 0 0 0 |
| − | 26 27 1 1 0 0 0 | + | 26 27 1 1 0 0 0 |
| − | 27 28 1 1 0 0 0 | + | 27 28 1 1 0 0 0 |
| − | 29 28 1 1 0 0 0 | + | 29 28 1 1 0 0 0 |
| − | 29 30 1 0 0 0 0 | + | 29 30 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 31 26 1 0 0 0 0 | + | 31 26 1 0 0 0 0 |
| − | 26 32 1 0 0 0 0 | + | 26 32 1 0 0 0 0 |
| − | 27 33 1 0 0 0 0 | + | 27 33 1 0 0 0 0 |
| − | 28 34 1 0 0 0 0 | + | 28 34 1 0 0 0 0 |
| − | 29 18 1 0 0 0 0 | + | 29 18 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 1 1 0 0 0 | + | 36 37 1 1 0 0 0 |
| − | 37 38 1 1 0 0 0 | + | 37 38 1 1 0 0 0 |
| − | 39 38 1 1 0 0 0 | + | 39 38 1 1 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 40 35 1 0 0 0 0 | + | 40 35 1 0 0 0 0 |
| − | 35 41 1 0 0 0 0 | + | 35 41 1 0 0 0 0 |
| − | 40 42 1 0 0 0 0 | + | 40 42 1 0 0 0 0 |
| − | 39 43 1 0 0 0 0 | + | 39 43 1 0 0 0 0 |
| − | 36 19 1 0 0 0 0 | + | 36 19 1 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 15 44 1 0 0 0 0 | + | 15 44 1 0 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 38 46 1 0 0 0 0 | + | 38 46 1 0 0 0 0 |
| − | 48 49 1 0 0 0 0 | + | 48 49 1 0 0 0 0 |
| − | 31 48 1 0 0 0 0 | + | 31 48 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 44 45 | + | M SAL 1 2 44 45 |
| − | M SBL 1 1 49 | + | M SBL 1 1 49 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 49 -0.7283 -0.4205 | + | M SBV 1 49 -0.7283 -0.4205 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 46 47 | + | M SAL 2 2 46 47 |
| − | M SBL 2 1 51 | + | M SBL 2 1 51 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SBV 2 51 -0.5992 -0.1154 | + | M SBV 2 51 -0.5992 -0.1154 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 48 49 | + | M SAL 3 2 48 49 |
| − | M SBL 3 1 53 | + | M SBL 3 1 53 |
| − | M SMT 3 ^CH2OH | + | M SMT 3 ^CH2OH |
| − | M SBV 3 53 0.6536 -0.5305 | + | M SBV 3 53 0.6536 -0.5305 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FABGI0006 | + | ID FL5FABGI0006 |
| − | FORMULA C33H40O16 | + | FORMULA C33H40O16 |
| − | EXACTMASS 692.2316352319999 | + | EXACTMASS 692.2316352319999 |
| − | AVERAGEMASS 692.6611 | + | AVERAGEMASS 692.6611 |
| − | SMILES O(C(C5O)OC(C(O)C5O)CO)c(c4CC=C(C)C)cc(c(c14)C(=O)C(OC(C3O)OC(C(C(O)3)O)CO)=C(c(c2)ccc(c2)OC)O1)O | + | SMILES O(C(C5O)OC(C(O)C5O)CO)c(c4CC=C(C)C)cc(c(c14)C(=O)C(OC(C3O)OC(C(C(O)3)O)CO)=C(c(c2)ccc(c2)OC)O1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
49 53 0 0 0 0 0 0 0 0999 V2000
-1.6518 -0.4123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6518 -1.2377 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9370 -1.6504 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2223 -1.2377 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2223 -0.4123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9370 0.0003 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4925 -1.6504 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2073 -1.2377 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2073 -0.4123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4925 0.0003 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4925 -2.2938 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9218 0.0001 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6503 -0.4204 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3788 0.0001 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3788 0.8413 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6503 1.2618 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9218 0.8413 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3663 0.0001 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7137 -1.7374 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9370 -2.4754 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9370 0.8254 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6515 1.2378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6515 2.0628 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9370 2.4754 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3660 2.4754 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.9680 0.0141 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4910 -0.6155 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8043 -0.3484 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1417 -0.3413 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6231 0.1404 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2239 -0.1767 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.5649 -0.3066 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.1093 -0.6325 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1825 -0.8040 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9431 -1.1421 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5566 -1.8116 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2999 -1.5991 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.0477 -1.8116 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4343 -1.1421 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6909 -1.3545 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3504 -1.2946 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1014 -0.8259 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.0345 -1.2592 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1070 1.2618 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.0249 0.7318 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.6468 -1.6962 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.5649 -2.2262 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.8775 0.3537 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8536 0.0794 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
8 19 1 0 0 0 0
3 20 1 0 0 0 0
6 21 1 0 0 0 0
21 22 1 0 0 0 0
22 23 2 0 0 0 0
23 24 1 0 0 0 0
23 25 1 0 0 0 0
26 27 1 1 0 0 0
27 28 1 1 0 0 0
29 28 1 1 0 0 0
29 30 1 0 0 0 0
30 31 1 0 0 0 0
31 26 1 0 0 0 0
26 32 1 0 0 0 0
27 33 1 0 0 0 0
28 34 1 0 0 0 0
29 18 1 0 0 0 0
35 36 1 0 0 0 0
36 37 1 1 0 0 0
37 38 1 1 0 0 0
39 38 1 1 0 0 0
39 40 1 0 0 0 0
40 35 1 0 0 0 0
35 41 1 0 0 0 0
40 42 1 0 0 0 0
39 43 1 0 0 0 0
36 19 1 0 0 0 0
44 45 1 0 0 0 0
15 44 1 0 0 0 0
46 47 1 0 0 0 0
38 46 1 0 0 0 0
48 49 1 0 0 0 0
31 48 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 44 45
M SBL 1 1 49
M SMT 1 OCH3
M SBV 1 49 -0.7283 -0.4205
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 46 47
M SBL 2 1 51
M SMT 2 CH2OH
M SBV 2 51 -0.5992 -0.1154
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 48 49
M SBL 3 1 53
M SMT 3 ^CH2OH
M SBV 3 53 0.6536 -0.5305
S SKP 5
ID FL5FABGI0006
FORMULA C33H40O16
EXACTMASS 692.2316352319999
AVERAGEMASS 692.6611
SMILES O(C(C5O)OC(C(O)C5O)CO)c(c4CC=C(C)C)cc(c(c14)C(=O)C(OC(C3O)OC(C(C(O)3)O)CO)=C(c(c2)ccc(c2)OC)O1)O
M END