Mol:FL5FAAGS0120
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 62 67 0 0 0 0 0 0 0 0999 V2000 | + | 62 67 0 0 0 0 0 0 0 0999 V2000 |
| − | 1.0705 5.2379 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0705 5.2379 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0705 4.4129 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0705 4.4129 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7850 4.0004 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7850 4.0004 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4995 4.4129 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4995 4.4129 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4995 5.2379 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4995 5.2379 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7850 5.6504 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7850 5.6504 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3561 4.0004 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3561 4.0004 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3584 4.4129 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3584 4.4129 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0729 4.0004 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0729 4.0004 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0729 3.1754 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0729 3.1754 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3584 2.7629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3584 2.7629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3561 3.1754 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3561 3.1754 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7874 4.4129 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7874 4.4129 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5018 4.0004 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5018 4.0004 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5018 3.1754 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5018 3.1754 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7874 2.7629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7874 2.7629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3584 2.0446 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3584 2.0446 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2163 4.4129 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2163 4.4129 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9624 2.8253 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9624 2.8253 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1002 5.5847 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1002 5.5847 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7874 2.0633 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7874 2.0633 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1930 0.5102 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1930 0.5102 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5283 0.1097 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5283 0.1097 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1051 0.6996 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1051 0.6996 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9020 0.9132 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9020 0.9132 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1807 1.3138 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1807 1.3138 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6038 0.7239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6038 0.7239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1680 1.1454 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1680 1.1454 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3859 1.1361 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3859 1.1361 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3587 -0.1525 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3587 -0.1525 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0104 0.1011 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0104 0.1011 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2872 0.0628 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2872 0.0628 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0591 1.1248 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0591 1.1248 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3836 1.6657 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3836 1.6657 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3905 0.5280 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3905 0.5280 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1133 0.5158 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1133 0.5158 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5224 -0.2211 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5224 -0.2211 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4530 1.0820 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4530 1.0820 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1523 -1.1830 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1523 -1.1830 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5089 -1.6994 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5089 -1.6994 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1592 -1.2153 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1592 -1.2153 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9807 -1.1392 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9807 -1.1392 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3373 -0.6227 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3373 -0.6227 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3308 -1.1067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3308 -1.1067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6893 -0.6178 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6893 -0.6178 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2368 -0.5335 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2368 -0.5335 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4274 -1.8082 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4274 -1.8082 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0208 -1.6206 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0208 -1.6206 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2313 -1.8737 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2313 -1.8737 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9961 -1.9914 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9961 -1.9914 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4321 -1.4482 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4321 -1.4482 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2476 -2.6382 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2476 -2.6382 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8842 -2.7363 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8842 -2.7363 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1019 -3.2961 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1019 -3.2961 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5855 -3.9395 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5855 -3.9395 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8845 -4.7084 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8845 -4.7084 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6999 -4.8339 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6999 -4.8339 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2163 -4.1906 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2163 -4.1906 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9173 -3.4216 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9173 -3.4216 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9334 -5.4344 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9334 -5.4344 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5969 -5.3619 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5969 -5.3619 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8550 -5.6504 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8550 -5.6504 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 2 7 1 0 0 0 0 | + | 2 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 11 1 0 0 0 0 | + | 10 11 1 0 0 0 0 |
| − | 11 12 1 0 0 0 0 | + | 11 12 1 0 0 0 0 |
| − | 12 7 2 0 0 0 0 | + | 12 7 2 0 0 0 0 |
| − | 9 13 1 0 0 0 0 | + | 9 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 10 1 0 0 0 0 | + | 16 10 1 0 0 0 0 |
| − | 14 18 1 0 0 0 0 | + | 14 18 1 0 0 0 0 |
| − | 11 17 2 0 0 0 0 | + | 11 17 2 0 0 0 0 |
| − | 12 19 1 0 0 0 0 | + | 12 19 1 0 0 0 0 |
| − | 5 20 1 0 0 0 0 | + | 5 20 1 0 0 0 0 |
| − | 16 21 1 0 0 0 0 | + | 16 21 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 22 30 1 0 0 0 0 | + | 22 30 1 0 0 0 0 |
| − | 23 31 1 0 0 0 0 | + | 23 31 1 0 0 0 0 |
| − | 24 32 1 0 0 0 0 | + | 24 32 1 0 0 0 0 |
| − | 29 33 1 0 0 0 0 | + | 29 33 1 0 0 0 0 |
| − | 33 34 2 0 0 0 0 | + | 33 34 2 0 0 0 0 |
| − | 33 35 1 0 0 0 0 | + | 33 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 2 0 0 0 0 | + | 36 37 2 0 0 0 0 |
| − | 36 38 1 0 0 0 0 | + | 36 38 1 0 0 0 0 |
| − | 39 40 1 1 0 0 0 | + | 39 40 1 1 0 0 0 |
| − | 40 41 1 1 0 0 0 | + | 40 41 1 1 0 0 0 |
| − | 42 41 1 1 0 0 0 | + | 42 41 1 1 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 44 39 1 0 0 0 0 | + | 44 39 1 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | 39 47 1 0 0 0 0 | + | 39 47 1 0 0 0 0 |
| − | 40 48 1 0 0 0 0 | + | 40 48 1 0 0 0 0 |
| − | 41 49 1 0 0 0 0 | + | 41 49 1 0 0 0 0 |
| − | 49 50 1 0 0 0 0 | + | 49 50 1 0 0 0 0 |
| − | 50 51 2 0 0 0 0 | + | 50 51 2 0 0 0 0 |
| − | 50 52 1 0 0 0 0 | + | 50 52 1 0 0 0 0 |
| − | 52 53 2 0 0 0 0 | + | 52 53 2 0 0 0 0 |
| − | 53 54 1 0 0 0 0 | + | 53 54 1 0 0 0 0 |
| − | 54 55 2 0 0 0 0 | + | 54 55 2 0 0 0 0 |
| − | 55 56 1 0 0 0 0 | + | 55 56 1 0 0 0 0 |
| − | 56 57 2 0 0 0 0 | + | 56 57 2 0 0 0 0 |
| − | 57 58 1 0 0 0 0 | + | 57 58 1 0 0 0 0 |
| − | 58 59 2 0 0 0 0 | + | 58 59 2 0 0 0 0 |
| − | 59 54 1 0 0 0 0 | + | 59 54 1 0 0 0 0 |
| − | 57 60 1 0 0 0 0 | + | 57 60 1 0 0 0 0 |
| − | 56 61 1 0 0 0 0 | + | 56 61 1 0 0 0 0 |
| − | 61 62 1 0 0 0 0 | + | 61 62 1 0 0 0 0 |
| − | 25 19 1 0 0 0 0 | + | 25 19 1 0 0 0 0 |
| − | 32 42 1 0 0 0 0 | + | 32 42 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FAAGS0120 | + | ID FL5FAAGS0120 |
| − | KNApSAcK_ID C00013787 | + | KNApSAcK_ID C00013787 |
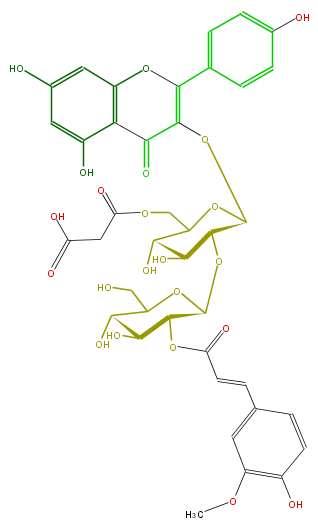
| − | NAME Kaempferol 3-(2''-feruloylglucosyl)-(1->2)-(6''-malonylglucoside) | + | NAME Kaempferol 3-(2''-feruloylglucosyl)-(1->2)-(6''-malonylglucoside) |
| − | CAS_RN 219745-02-5 | + | CAS_RN 219745-02-5 |
| − | FORMULA C40H40O22 | + | FORMULA C40H40O22 |
| − | EXACTMASS 872.201122964 | + | EXACTMASS 872.201122964 |
| − | AVERAGEMASS 872.7324 | + | AVERAGEMASS 872.7324 |
| − | SMILES c(c6)(cc(OC)c(O)c6)C=CC(=O)OC(C1O)C(OC(C(OC(=C(c(c5)ccc(c5)O)4)C(c(c(O4)3)c(cc(c3)O)O)=O)2)C(O)C(O)C(COC(=O)CC(O)=O)O2)OC(C(O)1)CO | + | SMILES c(c6)(cc(OC)c(O)c6)C=CC(=O)OC(C1O)C(OC(C(OC(=C(c(c5)ccc(c5)O)4)C(c(c(O4)3)c(cc(c3)O)O)=O)2)C(O)C(O)C(COC(=O)CC(O)=O)O2)OC(C(O)1)CO |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
62 67 0 0 0 0 0 0 0 0999 V2000
1.0705 5.2379 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0705 4.4129 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7850 4.0004 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4995 4.4129 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4995 5.2379 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7850 5.6504 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3561 4.0004 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3584 4.4129 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0729 4.0004 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0729 3.1754 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3584 2.7629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3561 3.1754 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7874 4.4129 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5018 4.0004 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5018 3.1754 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7874 2.7629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3584 2.0446 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2163 4.4129 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9624 2.8253 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1002 5.5847 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7874 2.0633 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1930 0.5102 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5283 0.1097 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1051 0.6996 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9020 0.9132 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1807 1.3138 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6038 0.7239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1680 1.1454 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3859 1.1361 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.3587 -0.1525 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0104 0.1011 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2872 0.0628 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0591 1.1248 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3836 1.6657 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3905 0.5280 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1133 0.5158 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5224 -0.2211 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4530 1.0820 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.1523 -1.1830 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5089 -1.6994 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1592 -1.2153 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9807 -1.1392 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3373 -0.6227 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.3308 -1.1067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6893 -0.6178 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2368 -0.5335 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4274 -1.8082 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0208 -1.6206 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2313 -1.8737 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9961 -1.9914 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4321 -1.4482 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2476 -2.6382 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8842 -2.7363 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1019 -3.2961 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5855 -3.9395 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8845 -4.7084 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6999 -4.8339 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2163 -4.1906 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9173 -3.4216 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9334 -5.4344 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5969 -5.3619 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8550 -5.6504 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
2 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 11 1 0 0 0 0
11 12 1 0 0 0 0
12 7 2 0 0 0 0
9 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 10 1 0 0 0 0
14 18 1 0 0 0 0
11 17 2 0 0 0 0
12 19 1 0 0 0 0
5 20 1 0 0 0 0
16 21 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
27 28 1 0 0 0 0
28 29 1 0 0 0 0
22 30 1 0 0 0 0
23 31 1 0 0 0 0
24 32 1 0 0 0 0
29 33 1 0 0 0 0
33 34 2 0 0 0 0
33 35 1 0 0 0 0
35 36 1 0 0 0 0
36 37 2 0 0 0 0
36 38 1 0 0 0 0
39 40 1 1 0 0 0
40 41 1 1 0 0 0
42 41 1 1 0 0 0
42 43 1 0 0 0 0
43 44 1 0 0 0 0
44 39 1 0 0 0 0
44 45 1 0 0 0 0
45 46 1 0 0 0 0
39 47 1 0 0 0 0
40 48 1 0 0 0 0
41 49 1 0 0 0 0
49 50 1 0 0 0 0
50 51 2 0 0 0 0
50 52 1 0 0 0 0
52 53 2 0 0 0 0
53 54 1 0 0 0 0
54 55 2 0 0 0 0
55 56 1 0 0 0 0
56 57 2 0 0 0 0
57 58 1 0 0 0 0
58 59 2 0 0 0 0
59 54 1 0 0 0 0
57 60 1 0 0 0 0
56 61 1 0 0 0 0
61 62 1 0 0 0 0
25 19 1 0 0 0 0
32 42 1 0 0 0 0
S SKP 8
ID FL5FAAGS0120
KNApSAcK_ID C00013787
NAME Kaempferol 3-(2''-feruloylglucosyl)-(1->2)-(6''-malonylglucoside)
CAS_RN 219745-02-5
FORMULA C40H40O22
EXACTMASS 872.201122964
AVERAGEMASS 872.7324
SMILES c(c6)(cc(OC)c(O)c6)C=CC(=O)OC(C1O)C(OC(C(OC(=C(c(c5)ccc(c5)O)4)C(c(c(O4)3)c(cc(c3)O)O)=O)2)C(O)C(O)C(COC(=O)CC(O)=O)O2)OC(C(O)1)CO
M END