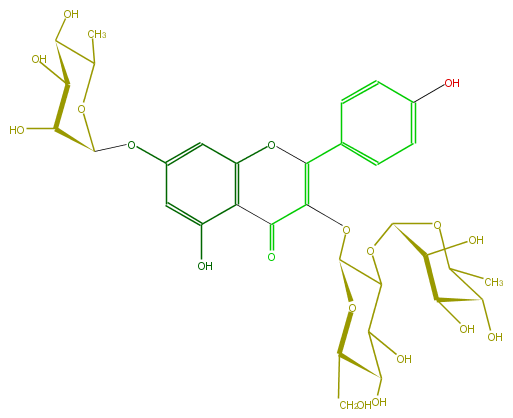
Mol:FL5FAAGL0043
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 52 57 0 0 0 0 0 0 0 0999 V2000 | + | 52 57 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.7954 0.8926 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7954 0.8926 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7954 0.0833 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7954 0.0833 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0944 -0.3215 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0944 -0.3215 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3934 0.0833 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3934 0.0833 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3934 0.8926 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3934 0.8926 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0944 1.2974 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0944 1.2974 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3076 -0.3215 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3076 -0.3215 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0086 0.0833 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0086 0.0833 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0086 0.8926 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0086 0.8926 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3076 1.2974 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3076 1.2974 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3076 -0.9526 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3076 -0.9526 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7094 1.2973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7094 1.2973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4239 0.8848 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4239 0.8848 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1384 1.2973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1384 1.2973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1384 2.1223 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1384 2.1223 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4239 2.5347 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4239 2.5347 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7094 2.1223 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7094 2.1223 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0944 -1.1306 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0944 -1.1306 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4962 1.2973 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4962 1.2973 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8527 2.5346 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8527 2.5346 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8161 -0.4082 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8161 -0.4082 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4991 -1.5156 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4991 -1.5156 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6530 -1.0271 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6530 -1.0271 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9215 -1.9665 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9215 -1.9665 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6530 -2.9116 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6530 -2.9116 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4991 -3.4002 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4991 -3.4002 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2307 -2.4607 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2307 -2.4607 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2987 -0.8366 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2987 -0.8366 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8814 -2.9932 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8814 -2.9932 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3888 -3.8473 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3888 -3.8473 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6044 -1.8296 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6044 -1.8296 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5187 -1.8296 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5187 -1.8296 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8616 -1.1940 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8616 -1.1940 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6371 -0.3026 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6371 -0.3026 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7228 -0.3026 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7228 -0.3026 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3799 -0.9382 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3799 -0.9382 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3250 -0.6192 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3250 -0.6192 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1600 -2.3851 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1600 -2.3851 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7096 -2.5425 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7096 -2.5425 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6418 -1.4429 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6418 -1.4429 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0216 1.5894 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0216 1.5894 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2298 1.1323 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2298 1.1323 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4811 2.0112 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4811 2.0112 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2298 2.8956 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2298 2.8956 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0216 3.3528 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0216 3.3528 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7704 2.4737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7704 2.4737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7791 3.9082 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7791 3.9082 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2932 3.5058 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2932 3.5058 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4193 3.0138 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4193 3.0138 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7096 1.5944 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7096 1.5944 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6404 -3.9082 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6404 -3.9082 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3806 -3.8812 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3806 -3.8812 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 8 21 1 0 0 0 0 | + | 8 21 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 27 29 1 0 0 0 0 | + | 27 29 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 23 21 1 0 0 0 0 | + | 23 21 1 0 0 0 0 |
| − | 32 31 1 1 0 0 0 | + | 32 31 1 1 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 1 0 0 0 | + | 35 36 1 1 0 0 0 |
| − | 36 31 1 1 0 0 0 | + | 36 31 1 1 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 31 38 1 0 0 0 0 | + | 31 38 1 0 0 0 0 |
| − | 32 39 1 0 0 0 0 | + | 32 39 1 0 0 0 0 |
| − | 33 40 1 0 0 0 0 | + | 33 40 1 0 0 0 0 |
| − | 35 28 1 0 0 0 0 | + | 35 28 1 0 0 0 0 |
| − | 42 41 1 1 0 0 0 | + | 42 41 1 1 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 45 46 1 1 0 0 0 | + | 45 46 1 1 0 0 0 |
| − | 46 41 1 1 0 0 0 | + | 46 41 1 1 0 0 0 |
| − | 45 47 1 0 0 0 0 | + | 45 47 1 0 0 0 0 |
| − | 44 48 1 0 0 0 0 | + | 44 48 1 0 0 0 0 |
| − | 46 49 1 0 0 0 0 | + | 46 49 1 0 0 0 0 |
| − | 41 50 1 0 0 0 0 | + | 41 50 1 0 0 0 0 |
| − | 42 19 1 0 0 0 0 | + | 42 19 1 0 0 0 0 |
| − | 51 52 1 0 0 0 0 | + | 51 52 1 0 0 0 0 |
| − | 25 51 1 0 0 0 0 | + | 25 51 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 51 52 | + | M SAL 1 2 51 52 |
| − | M SBL 1 1 57 | + | M SBL 1 1 57 |
| − | M SMT 1 ^ CH2OH | + | M SMT 1 ^ CH2OH |
| − | M SBV 1 57 0.0125 0.9966 | + | M SBV 1 57 0.0125 0.9966 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FAAGL0043 | + | ID FL5FAAGL0043 |
| − | FORMULA C33H40O19 | + | FORMULA C33H40O19 |
| − | EXACTMASS 740.216379098 | + | EXACTMASS 740.216379098 |
| − | AVERAGEMASS 740.6593 | + | AVERAGEMASS 740.6593 |
| − | SMILES C(O6)(C(C(O)C(O)C6CO)OC(O5)C(O)C(O)C(O)C5C)OC(C3=O)=C(c(c4)ccc(O)c4)Oc(c31)cc(OC(C2O)OC(C)C(C2O)O)cc1O | + | SMILES C(O6)(C(C(O)C(O)C6CO)OC(O5)C(O)C(O)C(O)C5C)OC(C3=O)=C(c(c4)ccc(O)c4)Oc(c31)cc(OC(C2O)OC(C)C(C2O)O)cc1O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
52 57 0 0 0 0 0 0 0 0999 V2000
-1.7954 0.8926 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7954 0.0833 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0944 -0.3215 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3934 0.0833 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3934 0.8926 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0944 1.2974 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3076 -0.3215 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0086 0.0833 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0086 0.8926 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3076 1.2974 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3076 -0.9526 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7094 1.2973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4239 0.8848 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1384 1.2973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1384 2.1223 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4239 2.5347 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7094 2.1223 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0944 -1.1306 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4962 1.2973 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.8527 2.5346 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8161 -0.4082 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4991 -1.5156 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6530 -1.0271 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9215 -1.9665 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6530 -2.9116 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4991 -3.4002 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2307 -2.4607 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2987 -0.8366 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8814 -2.9932 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3888 -3.8473 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6044 -1.8296 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5187 -1.8296 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8616 -1.1940 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6371 -0.3026 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7228 -0.3026 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3799 -0.9382 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3250 -0.6192 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1600 -2.3851 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.7096 -2.5425 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.6418 -1.4429 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0216 1.5894 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2298 1.1323 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4811 2.0112 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2298 2.8956 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0216 3.3528 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7704 2.4737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7791 3.9082 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2932 3.5058 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4193 3.0138 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.7096 1.5944 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6404 -3.9082 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3806 -3.8812 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
1 19 1 0 0 0 0
15 20 1 0 0 0 0
8 21 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
27 29 1 0 0 0 0
26 30 1 0 0 0 0
23 21 1 0 0 0 0
32 31 1 1 0 0 0
32 33 1 0 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 36 1 1 0 0 0
36 31 1 1 0 0 0
36 37 1 0 0 0 0
31 38 1 0 0 0 0
32 39 1 0 0 0 0
33 40 1 0 0 0 0
35 28 1 0 0 0 0
42 41 1 1 0 0 0
42 43 1 0 0 0 0
43 44 1 0 0 0 0
44 45 1 0 0 0 0
45 46 1 1 0 0 0
46 41 1 1 0 0 0
45 47 1 0 0 0 0
44 48 1 0 0 0 0
46 49 1 0 0 0 0
41 50 1 0 0 0 0
42 19 1 0 0 0 0
51 52 1 0 0 0 0
25 51 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 51 52
M SBL 1 1 57
M SMT 1 ^ CH2OH
M SBV 1 57 0.0125 0.9966
S SKP 5
ID FL5FAAGL0043
FORMULA C33H40O19
EXACTMASS 740.216379098
AVERAGEMASS 740.6593
SMILES C(O6)(C(C(O)C(O)C6CO)OC(O5)C(O)C(O)C(O)C5C)OC(C3=O)=C(c(c4)ccc(O)c4)Oc(c31)cc(OC(C2O)OC(C)C(C2O)O)cc1O
M END