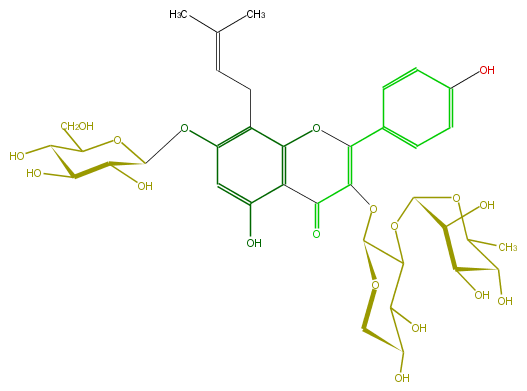
Mol:FL5FAAGI0009
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 56 61 0 0 0 0 0 0 0 0999 V2000 | + | 56 61 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.7378 1.0506 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7378 1.0506 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7378 0.2256 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7378 0.2256 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0234 -0.1868 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0234 -0.1868 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6911 0.2256 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6911 0.2256 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6911 1.0506 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6911 1.0506 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0234 1.4631 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0234 1.4631 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4055 -0.1868 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4055 -0.1868 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1199 0.2256 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1199 0.2256 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1199 1.0506 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1199 1.0506 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4055 1.4631 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4055 1.4631 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4055 -0.8301 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4055 -0.8301 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8341 1.4629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8341 1.4629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5622 1.0425 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5622 1.0425 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2903 1.4629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2903 1.4629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2903 2.3037 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2903 2.3037 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5622 2.7241 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5622 2.7241 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8341 2.3037 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8341 2.3037 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4520 1.4629 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4520 1.4629 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0183 2.7239 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0183 2.7239 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6262 -0.2738 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6262 -0.2738 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2675 -1.4692 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2675 -1.4692 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4053 -0.9714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4053 -0.9714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6790 -1.9287 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6790 -1.9287 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4053 -2.8919 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4053 -2.8919 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2675 -3.3898 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2675 -3.3898 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9941 -2.4324 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9941 -2.4324 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0830 -0.6492 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0830 -0.6492 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5338 -2.8486 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5338 -2.8486 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1864 -3.9370 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1864 -3.9370 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3941 -1.6000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3941 -1.6000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.3257 -1.6000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.3257 -1.6000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6561 -0.9523 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6561 -0.9523 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4272 -0.0439 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4272 -0.0439 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4956 -0.0438 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4956 -0.0438 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1651 -0.6917 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1651 -0.6917 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0219 -0.1970 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0219 -0.1970 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8998 -2.1470 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8998 -2.1470 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.3980 -2.2660 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.3980 -2.2660 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.4017 -1.1119 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.4017 -1.1119 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0234 2.2877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0234 2.2877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0234 -1.0116 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0234 -1.0116 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7375 2.7000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7375 2.7000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7375 3.5247 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7375 3.5247 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0234 3.9370 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0234 3.9370 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4517 3.9370 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4517 3.9370 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3627 1.0795 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3627 1.0795 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8421 0.3921 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8421 0.3921 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0923 0.6838 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0923 0.6838 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3109 0.6750 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3109 0.6750 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8946 1.2174 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8946 1.2174 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6605 0.9424 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6605 0.9424 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9896 0.8686 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9896 0.8686 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6209 0.4864 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6209 0.4864 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3725 0.2087 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3725 0.2087 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1487 1.5177 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1487 1.5177 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.4017 1.7990 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.4017 1.7990 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 15 19 1 0 0 0 0 | + | 15 19 1 0 0 0 0 |
| − | 8 20 1 0 0 0 0 | + | 8 20 1 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 26 28 1 0 0 0 0 | + | 26 28 1 0 0 0 0 |
| − | 25 29 1 0 0 0 0 | + | 25 29 1 0 0 0 0 |
| − | 22 20 1 0 0 0 0 | + | 22 20 1 0 0 0 0 |
| − | 31 30 1 1 0 0 0 | + | 31 30 1 1 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 1 0 0 0 | + | 34 35 1 1 0 0 0 |
| − | 35 30 1 1 0 0 0 | + | 35 30 1 1 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 30 37 1 0 0 0 0 | + | 30 37 1 0 0 0 0 |
| − | 31 38 1 0 0 0 0 | + | 31 38 1 0 0 0 0 |
| − | 32 39 1 0 0 0 0 | + | 32 39 1 0 0 0 0 |
| − | 34 27 1 0 0 0 0 | + | 34 27 1 0 0 0 0 |
| − | 6 40 1 0 0 0 0 | + | 6 40 1 0 0 0 0 |
| − | 3 41 1 0 0 0 0 | + | 3 41 1 0 0 0 0 |
| − | 40 42 1 0 0 0 0 | + | 40 42 1 0 0 0 0 |
| − | 42 43 2 0 0 0 0 | + | 42 43 2 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 43 45 1 0 0 0 0 | + | 43 45 1 0 0 0 0 |
| − | 46 47 1 1 0 0 0 | + | 46 47 1 1 0 0 0 |
| − | 47 48 1 1 0 0 0 | + | 47 48 1 1 0 0 0 |
| − | 49 48 1 1 0 0 0 | + | 49 48 1 1 0 0 0 |
| − | 49 50 1 0 0 0 0 | + | 49 50 1 0 0 0 0 |
| − | 50 51 1 0 0 0 0 | + | 50 51 1 0 0 0 0 |
| − | 51 46 1 0 0 0 0 | + | 51 46 1 0 0 0 0 |
| − | 46 52 1 0 0 0 0 | + | 46 52 1 0 0 0 0 |
| − | 47 53 1 0 0 0 0 | + | 47 53 1 0 0 0 0 |
| − | 48 54 1 0 0 0 0 | + | 48 54 1 0 0 0 0 |
| − | 49 18 1 0 0 0 0 | + | 49 18 1 0 0 0 0 |
| − | 55 56 1 0 0 0 0 | + | 55 56 1 0 0 0 0 |
| − | 51 55 1 0 0 0 0 | + | 51 55 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 55 56 | + | M SAL 1 2 55 56 |
| − | M SBL 1 1 61 | + | M SBL 1 1 61 |
| − | M SMT 1 ^ CH2OH | + | M SMT 1 ^ CH2OH |
| − | M SBV 1 61 0.4882 -0.5753 | + | M SBV 1 61 0.4882 -0.5753 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FAAGI0009 | + | ID FL5FAAGI0009 |
| − | FORMULA C37H46O19 | + | FORMULA C37H46O19 |
| − | EXACTMASS 794.26332929 | + | EXACTMASS 794.26332929 |
| − | AVERAGEMASS 794.74974 | + | AVERAGEMASS 794.74974 |
| − | SMILES OC(C(O)1)C(C(OC(C(OC(C(=O)4)=C(Oc(c5CC=C(C)C)c(c(cc5OC(C(O)6)OC(CO)C(C6O)O)O)4)c(c3)ccc(c3)O)2)C(O)C(CO2)O)OC1C)O | + | SMILES OC(C(O)1)C(C(OC(C(OC(C(=O)4)=C(Oc(c5CC=C(C)C)c(c(cc5OC(C(O)6)OC(CO)C(C6O)O)O)4)c(c3)ccc(c3)O)2)C(O)C(CO2)O)OC1C)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
56 61 0 0 0 0 0 0 0 0999 V2000
-0.7378 1.0506 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7378 0.2256 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0234 -0.1868 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6911 0.2256 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6911 1.0506 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0234 1.4631 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4055 -0.1868 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1199 0.2256 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1199 1.0506 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4055 1.4631 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4055 -0.8301 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8341 1.4629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5622 1.0425 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2903 1.4629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2903 2.3037 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5622 2.7241 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8341 2.3037 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4520 1.4629 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.0183 2.7239 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6262 -0.2738 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2675 -1.4692 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4053 -0.9714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6790 -1.9287 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4053 -2.8919 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2675 -3.3898 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9941 -2.4324 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0830 -0.6492 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5338 -2.8486 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1864 -3.9370 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.3941 -1.6000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.3257 -1.6000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.6561 -0.9523 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4272 -0.0439 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4956 -0.0438 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1651 -0.6917 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.0219 -0.1970 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.8998 -2.1470 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.3980 -2.2660 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.4017 -1.1119 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0234 2.2877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0234 -1.0116 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7375 2.7000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7375 3.5247 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0234 3.9370 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4517 3.9370 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3627 1.0795 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8421 0.3921 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0923 0.6838 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3109 0.6750 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8946 1.2174 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6605 0.9424 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.9896 0.8686 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.6209 0.4864 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3725 0.2087 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1487 1.5177 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.4017 1.7990 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
15 19 1 0 0 0 0
8 20 1 0 0 0 0
21 22 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
26 28 1 0 0 0 0
25 29 1 0 0 0 0
22 20 1 0 0 0 0
31 30 1 1 0 0 0
31 32 1 0 0 0 0
32 33 1 0 0 0 0
33 34 1 0 0 0 0
34 35 1 1 0 0 0
35 30 1 1 0 0 0
35 36 1 0 0 0 0
30 37 1 0 0 0 0
31 38 1 0 0 0 0
32 39 1 0 0 0 0
34 27 1 0 0 0 0
6 40 1 0 0 0 0
3 41 1 0 0 0 0
40 42 1 0 0 0 0
42 43 2 0 0 0 0
43 44 1 0 0 0 0
43 45 1 0 0 0 0
46 47 1 1 0 0 0
47 48 1 1 0 0 0
49 48 1 1 0 0 0
49 50 1 0 0 0 0
50 51 1 0 0 0 0
51 46 1 0 0 0 0
46 52 1 0 0 0 0
47 53 1 0 0 0 0
48 54 1 0 0 0 0
49 18 1 0 0 0 0
55 56 1 0 0 0 0
51 55 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 55 56
M SBL 1 1 61
M SMT 1 ^ CH2OH
M SBV 1 61 0.4882 -0.5753
S SKP 5
ID FL5FAAGI0009
FORMULA C37H46O19
EXACTMASS 794.26332929
AVERAGEMASS 794.74974
SMILES OC(C(O)1)C(C(OC(C(OC(C(=O)4)=C(Oc(c5CC=C(C)C)c(c(cc5OC(C(O)6)OC(CO)C(C6O)O)O)4)c(c3)ccc(c3)O)2)C(O)C(CO2)O)OC1C)O
M END