Mol:FL5FAAGA0032
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 55 60 0 0 0 0 0 0 0 0999 V2000 | + | 55 60 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.6863 2.4647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6863 2.4647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6863 1.8223 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6863 1.8223 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1300 1.5011 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1300 1.5011 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5737 1.8223 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5737 1.8223 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5737 2.4647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5737 2.4647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1300 2.7858 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1300 2.7858 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0174 1.5011 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0174 1.5011 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4611 1.8223 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4611 1.8223 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4611 2.4647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4611 2.4647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0174 2.7858 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0174 2.7858 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0174 1.0003 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0174 1.0003 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0950 2.7857 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0950 2.7857 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6620 2.4584 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6620 2.4584 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2290 2.7857 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2290 2.7857 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2290 3.4404 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2290 3.4404 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6620 3.7678 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6620 3.7678 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0950 3.4404 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0950 3.4404 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2424 2.7857 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2424 2.7857 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7958 3.7677 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7958 3.7677 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0669 1.4334 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0669 1.4334 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1300 0.8590 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1300 0.8590 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1966 0.0762 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1966 0.0762 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6896 0.4639 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6896 0.4639 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8208 -0.2172 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8208 -0.2172 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6461 -0.8321 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6461 -0.8321 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1966 -1.1501 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1966 -1.1501 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0220 -0.5388 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0220 -0.5388 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4579 -0.7905 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4579 -0.7905 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5336 -1.6916 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5336 -1.6916 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9510 1.7695 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9510 1.7695 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3527 1.2392 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3527 1.2392 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9312 1.4642 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9312 1.4642 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4893 1.4702 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4893 1.4702 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0837 1.8759 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0837 1.8759 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5777 1.6088 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5777 1.6088 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7265 1.2087 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7265 1.2087 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2626 0.9078 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2626 0.9078 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7152 1.0790 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7152 1.0790 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0655 0.5656 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0655 0.5656 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5336 -2.3219 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5336 -2.3219 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2647 -2.7877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2647 -2.7877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4132 -3.3417 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4132 -3.3417 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2320 -2.7877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2320 -2.7877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5026 -3.2564 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5026 -3.2564 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9746 -3.2564 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9746 -3.2564 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2698 -2.7451 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2698 -2.7451 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8602 -2.7451 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8602 -2.7451 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1554 -3.2564 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1554 -3.2564 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8602 -3.7678 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8602 -3.7678 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2698 -3.7678 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2698 -3.7678 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1552 -2.2343 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1552 -2.2343 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7453 -3.2564 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7453 -3.2564 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0119 -1.4700 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0119 -1.4700 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5279 1.8844 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5279 1.8844 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2424 1.4719 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2424 1.4719 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 15 19 1 0 0 0 0 | + | 15 19 1 0 0 0 0 |
| − | 8 20 1 0 0 0 0 | + | 8 20 1 0 0 0 0 |
| − | 3 21 1 0 0 0 0 | + | 3 21 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 33 32 1 1 0 0 0 | + | 33 32 1 1 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 30 1 0 0 0 0 | + | 35 30 1 0 0 0 0 |
| − | 31 36 1 0 0 0 0 | + | 31 36 1 0 0 0 0 |
| − | 32 37 1 0 0 0 0 | + | 32 37 1 0 0 0 0 |
| − | 33 38 1 0 0 0 0 | + | 33 38 1 0 0 0 0 |
| − | 30 20 1 0 0 0 0 | + | 30 20 1 0 0 0 0 |
| − | 23 36 1 0 0 0 0 | + | 23 36 1 0 0 0 0 |
| − | 22 39 1 0 0 0 0 | + | 22 39 1 0 0 0 0 |
| − | 29 40 1 0 0 0 0 | + | 29 40 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 41 42 2 0 0 0 0 | + | 41 42 2 0 0 0 0 |
| − | 41 43 1 0 0 0 0 | + | 41 43 1 0 0 0 0 |
| − | 43 44 2 0 0 0 0 | + | 43 44 2 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 45 46 2 0 0 0 0 | + | 45 46 2 0 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 47 48 2 0 0 0 0 | + | 47 48 2 0 0 0 0 |
| − | 48 49 1 0 0 0 0 | + | 48 49 1 0 0 0 0 |
| − | 49 50 2 0 0 0 0 | + | 49 50 2 0 0 0 0 |
| − | 50 45 1 0 0 0 0 | + | 50 45 1 0 0 0 0 |
| − | 47 51 1 0 0 0 0 | + | 47 51 1 0 0 0 0 |
| − | 48 52 1 0 0 0 0 | + | 48 52 1 0 0 0 0 |
| − | 25 29 1 0 0 0 0 | + | 25 29 1 0 0 0 0 |
| − | 26 53 1 0 0 0 0 | + | 26 53 1 0 0 0 0 |
| − | 34 54 1 0 0 0 0 | + | 34 54 1 0 0 0 0 |
| − | 54 55 1 0 0 0 0 | + | 54 55 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 54 55 | + | M SAL 1 2 54 55 |
| − | M SBL 1 1 59 | + | M SBL 1 1 59 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 59 -6.8580 6.3300 | + | M SBV 1 59 -6.8580 6.3300 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FAAGA0032 | + | ID FL5FAAGA0032 |
| − | KNApSAcK_ID C00005875 | + | KNApSAcK_ID C00005875 |
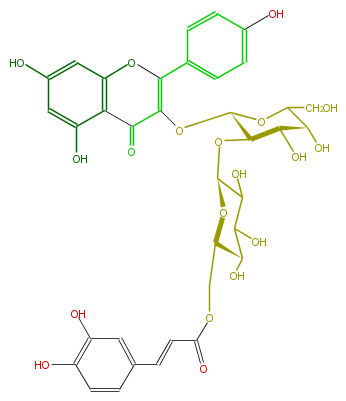
| − | NAME Kaempferol 3-(6'''-caffeylglucosyl)-(1->2)-galactoside | + | NAME Kaempferol 3-(6'''-caffeylglucosyl)-(1->2)-galactoside |
| − | CAS_RN 107140-37-4 | + | CAS_RN 107140-37-4 |
| − | FORMULA C36H36O19 | + | FORMULA C36H36O19 |
| − | EXACTMASS 772.18507897 | + | EXACTMASS 772.18507897 |
| − | AVERAGEMASS 772.6596400000001 | + | AVERAGEMASS 772.6596400000001 |
| − | SMILES C(OC(C(O)6)C(OC(C6O)CO)OC(C3=O)=C(c(c5)ccc(O)c5)Oc(c4)c3c(cc4O)O)(C(O)2)OC(C(C(O)2)O)COC(C=Cc(c1)cc(O)c(c1)O)=O | + | SMILES C(OC(C(O)6)C(OC(C6O)CO)OC(C3=O)=C(c(c5)ccc(O)c5)Oc(c4)c3c(cc4O)O)(C(O)2)OC(C(C(O)2)O)COC(C=Cc(c1)cc(O)c(c1)O)=O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
55 60 0 0 0 0 0 0 0 0999 V2000
-2.6863 2.4647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6863 1.8223 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1300 1.5011 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5737 1.8223 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5737 2.4647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1300 2.7858 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0174 1.5011 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4611 1.8223 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4611 2.4647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0174 2.7858 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0174 1.0003 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0950 2.7857 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6620 2.4584 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2290 2.7857 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2290 3.4404 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6620 3.7678 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0950 3.4404 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2424 2.7857 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7958 3.7677 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0669 1.4334 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1300 0.8590 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1966 0.0762 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6896 0.4639 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8208 -0.2172 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6461 -0.8321 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1966 -1.1501 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0220 -0.5388 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4579 -0.7905 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5336 -1.6916 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9510 1.7695 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3527 1.2392 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9312 1.4642 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4893 1.4702 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0837 1.8759 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5777 1.6088 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7265 1.2087 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2626 0.9078 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7152 1.0790 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0655 0.5656 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5336 -2.3219 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2647 -2.7877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4132 -3.3417 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2320 -2.7877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5026 -3.2564 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9746 -3.2564 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2698 -2.7451 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8602 -2.7451 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1554 -3.2564 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8602 -3.7678 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2698 -3.7678 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1552 -2.2343 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7453 -3.2564 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0119 -1.4700 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5279 1.8844 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2424 1.4719 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
15 19 1 0 0 0 0
8 20 1 0 0 0 0
3 21 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
27 28 1 0 0 0 0
30 31 1 1 0 0 0
31 32 1 1 0 0 0
33 32 1 1 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 30 1 0 0 0 0
31 36 1 0 0 0 0
32 37 1 0 0 0 0
33 38 1 0 0 0 0
30 20 1 0 0 0 0
23 36 1 0 0 0 0
22 39 1 0 0 0 0
29 40 1 0 0 0 0
40 41 1 0 0 0 0
41 42 2 0 0 0 0
41 43 1 0 0 0 0
43 44 2 0 0 0 0
44 45 1 0 0 0 0
45 46 2 0 0 0 0
46 47 1 0 0 0 0
47 48 2 0 0 0 0
48 49 1 0 0 0 0
49 50 2 0 0 0 0
50 45 1 0 0 0 0
47 51 1 0 0 0 0
48 52 1 0 0 0 0
25 29 1 0 0 0 0
26 53 1 0 0 0 0
34 54 1 0 0 0 0
54 55 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 54 55
M SBL 1 1 59
M SMT 1 CH2OH
M SBV 1 59 -6.8580 6.3300
S SKP 8
ID FL5FAAGA0032
KNApSAcK_ID C00005875
NAME Kaempferol 3-(6'''-caffeylglucosyl)-(1->2)-galactoside
CAS_RN 107140-37-4
FORMULA C36H36O19
EXACTMASS 772.18507897
AVERAGEMASS 772.6596400000001
SMILES C(OC(C(O)6)C(OC(C6O)CO)OC(C3=O)=C(c(c5)ccc(O)c5)Oc(c4)c3c(cc4O)O)(C(O)2)OC(C(C(O)2)O)COC(C=Cc(c1)cc(O)c(c1)O)=O
M END