Mol:FL5FAAGA0021
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 53 58 0 0 0 0 0 0 0 0999 V2000 | + | 53 58 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.0369 1.2551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0369 1.2551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0369 0.6127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0369 0.6127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4806 0.2915 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4806 0.2915 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0757 0.6127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0757 0.6127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0757 1.2551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0757 1.2551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4806 1.5763 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4806 1.5763 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6320 0.2915 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6320 0.2915 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1883 0.6127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1883 0.6127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1883 1.2551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1883 1.2551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6320 1.5763 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6320 1.5763 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6320 -0.2093 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6320 -0.2093 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7444 1.5762 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7444 1.5762 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3114 1.2488 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3114 1.2488 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8784 1.5762 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8784 1.5762 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8784 2.2308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8784 2.2308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3114 2.5582 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3114 2.5582 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7444 2.2308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7444 2.2308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4806 -0.3506 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4806 -0.3506 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5930 1.5762 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5930 1.5762 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4452 2.5581 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4452 2.5581 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5825 0.2238 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5825 0.2238 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3711 -0.8033 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 2.3711 -0.8033 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 1.6996 -0.4157 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.6996 -0.4157 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.9127 -1.1611 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9127 -1.1611 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6996 -1.9111 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.6996 -1.9111 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.3711 -2.2989 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.3711 -2.2989 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.1581 -1.5533 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 2.1581 -1.5533 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 2.1781 -0.0834 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1781 -0.0834 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7696 -1.6069 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7696 -1.6069 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5737 -2.8554 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5737 -2.8554 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2482 -0.9052 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 3.2482 -0.9052 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 3.9737 -0.9052 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 3.9737 -0.9052 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.4523 -0.4009 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 3.4523 -0.4009 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 3.2740 0.3065 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2740 0.3065 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5486 0.3066 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.5486 0.3066 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.0699 -0.1979 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 3.0699 -0.1979 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.7371 0.1873 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7371 0.1873 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6891 -1.3461 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6891 -1.3461 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1252 -1.4709 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1252 -1.4709 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1685 -0.4009 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1685 -0.4009 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7151 1.2157 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -3.7151 1.2157 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -3.3097 0.6805 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -3.3097 0.6805 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.7259 0.9076 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.7259 0.9076 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.1174 0.9009 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.1174 0.9009 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.5719 1.3231 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5719 1.3231 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1683 1.1090 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -3.1683 1.1090 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -4.2365 1.2614 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2365 1.2614 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6924 0.1768 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6924 0.1768 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3914 0.3460 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3914 0.3460 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3128 -1.9265 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3128 -1.9265 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5983 -2.3388 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5983 -2.3388 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3889 1.8150 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3889 1.8150 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3646 2.0340 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3646 2.0340 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 8 21 1 0 0 0 0 | + | 8 21 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 27 29 1 0 0 0 0 | + | 27 29 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 23 21 1 0 0 0 0 | + | 23 21 1 0 0 0 0 |
| − | 32 31 1 1 0 0 0 | + | 32 31 1 1 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 1 0 0 0 | + | 35 36 1 1 0 0 0 |
| − | 36 31 1 1 0 0 0 | + | 36 31 1 1 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 31 38 1 0 0 0 0 | + | 31 38 1 0 0 0 0 |
| − | 32 39 1 0 0 0 0 | + | 32 39 1 0 0 0 0 |
| − | 33 40 1 0 0 0 0 | + | 33 40 1 0 0 0 0 |
| − | 35 28 1 0 0 0 0 | + | 35 28 1 0 0 0 0 |
| − | 41 42 1 1 0 0 0 | + | 41 42 1 1 0 0 0 |
| − | 42 43 1 1 0 0 0 | + | 42 43 1 1 0 0 0 |
| − | 44 43 1 1 0 0 0 | + | 44 43 1 1 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | 46 41 1 0 0 0 0 | + | 46 41 1 0 0 0 0 |
| − | 41 47 1 0 0 0 0 | + | 41 47 1 0 0 0 0 |
| − | 42 48 1 0 0 0 0 | + | 42 48 1 0 0 0 0 |
| − | 43 49 1 0 0 0 0 | + | 43 49 1 0 0 0 0 |
| − | 44 19 1 0 0 0 0 | + | 44 19 1 0 0 0 0 |
| − | 25 50 1 0 0 0 0 | + | 25 50 1 0 0 0 0 |
| − | 50 51 1 0 0 0 0 | + | 50 51 1 0 0 0 0 |
| − | 46 52 1 0 0 0 0 | + | 46 52 1 0 0 0 0 |
| − | 52 53 1 0 0 0 0 | + | 52 53 1 0 0 0 0 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 52 53 | + | M SAL 2 2 52 53 |
| − | M SBL 2 1 57 | + | M SBL 2 1 57 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SVB 2 57 -3.3889 1.815 | + | M SVB 2 57 -3.3889 1.815 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 50 51 | + | M SAL 1 2 50 51 |
| − | M SBL 1 1 55 | + | M SBL 1 1 55 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SVB 1 55 1.3915 -2.1457 | + | M SVB 1 55 1.3915 -2.1457 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FAAGA0021 | + | ID FL5FAAGA0021 |
| − | KNApSAcK_ID C00005223 | + | KNApSAcK_ID C00005223 |
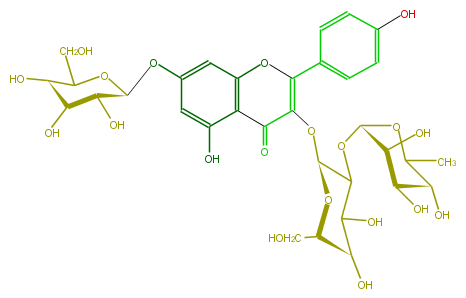
| − | NAME Kaempferol 3-rhamnosyl-(1->2)-galactoside-7-glucoside | + | NAME Kaempferol 3-rhamnosyl-(1->2)-galactoside-7-glucoside |
| − | CAS_RN 128988-58-9 | + | CAS_RN 128988-58-9 |
| − | FORMULA C33H40O20 | + | FORMULA C33H40O20 |
| − | EXACTMASS 756.21129372 | + | EXACTMASS 756.21129372 |
| − | AVERAGEMASS 756.6587 | + | AVERAGEMASS 756.6587 |
| − | SMILES Oc(c1)ccc(C(O2)=C(O[C@H](O6)C(C(O)[C@@H](O)[C@H]6CO)O[C@@H](O5)[C@@H](O)[C@H](O)[C@H](O)C5C)C(c(c3O)c2cc(O[C@@H]([C@@H](O)4)OC(CO)[C@H](O)[C@@H]4O)c3)=O)c1 | + | SMILES Oc(c1)ccc(C(O2)=C(O[C@H](O6)C(C(O)[C@@H](O)[C@H]6CO)O[C@@H](O5)[C@@H](O)[C@H](O)[C@H](O)C5C)C(c(c3O)c2cc(O[C@@H]([C@@H](O)4)OC(CO)[C@H](O)[C@@H]4O)c3)=O)c1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
53 58 0 0 0 0 0 0 0 0999 V2000
-1.0369 1.2551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0369 0.6127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4806 0.2915 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0757 0.6127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0757 1.2551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4806 1.5763 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6320 0.2915 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1883 0.6127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1883 1.2551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6320 1.5763 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6320 -0.2093 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7444 1.5762 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3114 1.2488 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8784 1.5762 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8784 2.2308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3114 2.5582 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7444 2.2308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4806 -0.3506 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.5930 1.5762 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4452 2.5581 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5825 0.2238 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3711 -0.8033 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
1.6996 -0.4157 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.9127 -1.1611 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6996 -1.9111 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.3711 -2.2989 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.1581 -1.5533 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
2.1781 -0.0834 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7696 -1.6069 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5737 -2.8554 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2482 -0.9052 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
3.9737 -0.9052 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.4523 -0.4009 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
3.2740 0.3065 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5486 0.3066 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.0699 -0.1979 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.7371 0.1873 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6891 -1.3461 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1252 -1.4709 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1685 -0.4009 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7151 1.2157 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-3.3097 0.6805 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.7259 0.9076 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.1174 0.9009 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.5719 1.3231 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1683 1.1090 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-4.2365 1.2614 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6924 0.1768 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3914 0.3460 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3128 -1.9265 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5983 -2.3388 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3889 1.8150 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3646 2.0340 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
1 19 1 0 0 0 0
15 20 1 0 0 0 0
8 21 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
27 29 1 0 0 0 0
26 30 1 0 0 0 0
23 21 1 0 0 0 0
32 31 1 1 0 0 0
32 33 1 0 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 36 1 1 0 0 0
36 31 1 1 0 0 0
36 37 1 0 0 0 0
31 38 1 0 0 0 0
32 39 1 0 0 0 0
33 40 1 0 0 0 0
35 28 1 0 0 0 0
41 42 1 1 0 0 0
42 43 1 1 0 0 0
44 43 1 1 0 0 0
44 45 1 0 0 0 0
45 46 1 0 0 0 0
46 41 1 0 0 0 0
41 47 1 0 0 0 0
42 48 1 0 0 0 0
43 49 1 0 0 0 0
44 19 1 0 0 0 0
25 50 1 0 0 0 0
50 51 1 0 0 0 0
46 52 1 0 0 0 0
52 53 1 0 0 0 0
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 52 53
M SBL 2 1 57
M SMT 2 CH2OH
M SVB 2 57 -3.3889 1.815
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 50 51
M SBL 1 1 55
M SMT 1 CH2OH
M SVB 1 55 1.3915 -2.1457
S SKP 8
ID FL5FAAGA0021
KNApSAcK_ID C00005223
NAME Kaempferol 3-rhamnosyl-(1->2)-galactoside-7-glucoside
CAS_RN 128988-58-9
FORMULA C33H40O20
EXACTMASS 756.21129372
AVERAGEMASS 756.6587
SMILES Oc(c1)ccc(C(O2)=C(O[C@H](O6)C(C(O)[C@@H](O)[C@H]6CO)O[C@@H](O5)[C@@H](O)[C@H](O)[C@H](O)C5C)C(c(c3O)c2cc(O[C@@H]([C@@H](O)4)OC(CO)[C@H](O)[C@@H]4O)c3)=O)c1
M END