Mol:FL5FAAGA0006
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 43 47 0 0 0 0 0 0 0 0999 V2000 | + | 43 47 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.4359 1.2052 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4359 1.2052 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4359 0.5628 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4359 0.5628 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8796 0.2416 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8796 0.2416 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3233 0.5628 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3233 0.5628 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3233 1.2052 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3233 1.2052 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8796 1.5264 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8796 1.5264 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7670 0.2416 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7670 0.2416 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2107 0.5628 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2107 0.5628 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2107 1.2052 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2107 1.2052 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7670 1.5264 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7670 1.5264 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7670 -0.2592 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7670 -0.2592 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3454 1.5262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3454 1.5262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9124 1.1989 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9124 1.1989 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4793 1.5262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4793 1.5262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4793 2.1809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4793 2.1809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9124 2.5083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9124 2.5083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3454 2.1809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3454 2.1809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8796 -0.4005 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8796 -0.4005 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9920 1.5262 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9920 1.5262 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0461 2.5082 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0461 2.5082 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1735 0.2977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1735 0.2977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7858 -0.3738 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7858 -0.3738 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5313 -0.1607 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5313 -0.1607 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2812 -0.3738 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2812 -0.3738 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6690 0.2977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6690 0.2977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9235 0.0847 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9235 0.0847 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4535 0.1048 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4535 0.1048 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2304 0.6163 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2304 0.6163 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4727 1.0303 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4727 1.0303 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9920 -0.3738 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9920 -0.3738 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8631 -0.9516 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8631 -0.9516 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3509 -1.4022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3509 -1.4022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8665 -2.0828 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8665 -2.0828 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6090 -1.7941 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6090 -1.7941 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3255 -1.7863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3255 -1.7863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8049 -1.2656 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8049 -1.2656 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0464 -1.5380 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0464 -1.5380 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2256 -1.7350 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2256 -1.7350 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0627 -2.1220 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0627 -2.1220 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0345 -2.5083 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0345 -2.5083 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8491 0.2108 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8491 0.2108 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7509 -0.7305 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7509 -0.7305 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4653 -1.1430 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4653 -1.1430 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 26 28 1 0 0 0 0 | + | 26 28 1 0 0 0 0 |
| − | 25 29 1 0 0 0 0 | + | 25 29 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 35 34 1 1 0 0 0 | + | 35 34 1 1 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 34 40 1 0 0 0 0 | + | 34 40 1 0 0 0 0 |
| − | 35 31 1 0 0 0 0 | + | 35 31 1 0 0 0 0 |
| − | 27 8 1 0 0 0 0 | + | 27 8 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 21 41 1 0 0 0 0 | + | 21 41 1 0 0 0 0 |
| − | 37 42 1 0 0 0 0 | + | 37 42 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 42 43 | + | M SAL 1 2 42 43 |
| − | M SBL 1 1 46 | + | M SBL 1 1 46 |
| − | M SMT 1 ^CH2OH | + | M SMT 1 ^CH2OH |
| − | M SBV 1 46 -5.7386 4.9207 | + | M SBV 1 46 -5.7386 4.9207 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FAAGA0006 | + | ID FL5FAAGA0006 |
| − | KNApSAcK_ID C00005158 | + | KNApSAcK_ID C00005158 |
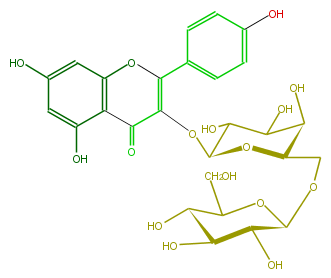
| − | NAME Kaempferol 3-glucosyl-(1->6)-galactoside | + | NAME Kaempferol 3-glucosyl-(1->6)-galactoside |
| − | CAS_RN 73803-52-8 | + | CAS_RN 73803-52-8 |
| − | FORMULA C27H30O16 | + | FORMULA C27H30O16 |
| − | EXACTMASS 610.153384912 | + | EXACTMASS 610.153384912 |
| − | AVERAGEMASS 610.5175 | + | AVERAGEMASS 610.5175 |
| − | SMILES c(O)(c1)cc(c(C(=O)2)c(OC(c(c5)ccc(c5)O)=C2OC(C3O)OC(COC(C4O)OC(C(C(O)4)O)CO)C(C(O)3)O)1)O | + | SMILES c(O)(c1)cc(c(C(=O)2)c(OC(c(c5)ccc(c5)O)=C2OC(C3O)OC(COC(C4O)OC(C(C(O)4)O)CO)C(C(O)3)O)1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
43 47 0 0 0 0 0 0 0 0999 V2000
-2.4359 1.2052 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4359 0.5628 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8796 0.2416 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3233 0.5628 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3233 1.2052 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8796 1.5264 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7670 0.2416 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2107 0.5628 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2107 1.2052 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7670 1.5264 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7670 -0.2592 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3454 1.5262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9124 1.1989 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4793 1.5262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4793 2.1809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9124 2.5083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3454 2.1809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8796 -0.4005 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9920 1.5262 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0461 2.5082 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1735 0.2977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7858 -0.3738 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5313 -0.1607 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2812 -0.3738 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6690 0.2977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9235 0.0847 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4535 0.1048 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2304 0.6163 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4727 1.0303 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9920 -0.3738 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8631 -0.9516 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3509 -1.4022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8665 -2.0828 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6090 -1.7941 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3255 -1.7863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8049 -1.2656 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0464 -1.5380 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2256 -1.7350 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0627 -2.1220 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0345 -2.5083 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8491 0.2108 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7509 -0.7305 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4653 -1.1430 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
1 19 1 0 0 0 0
15 20 1 0 0 0 0
21 22 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
26 28 1 0 0 0 0
25 29 1 0 0 0 0
24 30 1 0 0 0 0
30 31 1 0 0 0 0
32 33 1 1 0 0 0
33 34 1 1 0 0 0
35 34 1 1 0 0 0
35 36 1 0 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
34 40 1 0 0 0 0
35 31 1 0 0 0 0
27 8 1 0 0 0 0
27 22 1 0 0 0 0
21 41 1 0 0 0 0
37 42 1 0 0 0 0
42 43 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 42 43
M SBL 1 1 46
M SMT 1 ^CH2OH
M SBV 1 46 -5.7386 4.9207
S SKP 8
ID FL5FAAGA0006
KNApSAcK_ID C00005158
NAME Kaempferol 3-glucosyl-(1->6)-galactoside
CAS_RN 73803-52-8
FORMULA C27H30O16
EXACTMASS 610.153384912
AVERAGEMASS 610.5175
SMILES c(O)(c1)cc(c(C(=O)2)c(OC(c(c5)ccc(c5)O)=C2OC(C3O)OC(COC(C4O)OC(C(C(O)4)O)CO)C(C(O)3)O)1)O
M END