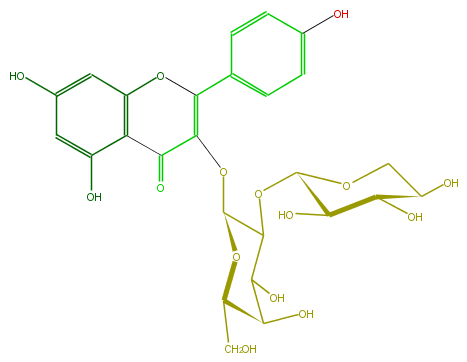
Mol:FL5FAAGA0004
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 41 45 0 0 0 0 0 0 0 0999 V2000 | + | 41 45 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.5706 1.9717 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5706 1.9717 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5706 1.1621 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5706 1.1621 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8695 0.7574 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8695 0.7574 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1685 1.1621 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1685 1.1621 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1685 1.9717 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1685 1.9717 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8695 2.3764 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8695 2.3764 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4674 0.7574 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4674 0.7574 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7664 1.1621 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7664 1.1621 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7664 1.9717 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7664 1.9717 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4674 2.3764 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4674 2.3764 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4674 0.1262 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4674 0.1262 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0655 2.3763 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0655 2.3763 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6490 1.9637 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6490 1.9637 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3634 2.3763 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3634 2.3763 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3634 3.2014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3634 3.2014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6490 3.6138 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6490 3.6138 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0655 3.2014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0655 3.2014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8695 -0.0519 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8695 -0.0519 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2714 2.3763 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2714 2.3763 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0777 3.6137 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0777 3.6137 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2114 -2.1359 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2114 -2.1359 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5803 -2.5930 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5803 -2.5930 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3290 -1.7140 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3290 -1.7140 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5803 -0.8297 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5803 -0.8297 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2114 -0.3725 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2114 -0.3725 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0397 -1.2516 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0397 -1.2516 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2114 0.4440 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2114 0.4440 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4809 0.0126 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4809 0.0126 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7889 -2.0721 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7889 -2.0721 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2413 0.4283 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2413 0.4283 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8910 -0.4294 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8910 -0.4294 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8267 -0.0656 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8267 -0.0656 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7297 -0.0559 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7297 -0.0559 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0736 0.6004 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0736 0.6004 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2550 0.1683 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2550 0.1683 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1063 -0.3977 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1063 -0.3977 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5513 -0.4528 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5513 -0.4528 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2714 0.2311 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2714 0.2311 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3769 -2.3824 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3769 -2.3824 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1206 -3.0940 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1206 -3.0940 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7798 -3.6138 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7798 -3.6138 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 22 21 1 1 0 0 0 | + | 22 21 1 1 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 1 0 0 0 | + | 25 26 1 1 0 0 0 |
| − | 26 21 1 1 0 0 0 | + | 26 21 1 1 0 0 0 |
| − | 25 27 1 0 0 0 0 | + | 25 27 1 0 0 0 0 |
| − | 24 28 1 0 0 0 0 | + | 24 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 33 32 1 1 0 0 0 | + | 33 32 1 1 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 30 1 0 0 0 0 | + | 35 30 1 0 0 0 0 |
| − | 31 36 1 0 0 0 0 | + | 31 36 1 0 0 0 0 |
| − | 32 37 1 0 0 0 0 | + | 32 37 1 0 0 0 0 |
| − | 30 28 1 0 0 0 0 | + | 30 28 1 0 0 0 0 |
| − | 33 38 1 0 0 0 0 | + | 33 38 1 0 0 0 0 |
| − | 22 39 1 0 0 0 0 | + | 22 39 1 0 0 0 0 |
| − | 27 8 1 0 0 0 0 | + | 27 8 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 21 40 1 0 0 0 0 | + | 21 40 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 40 41 | + | M SAL 1 2 40 41 |
| − | M SBL 1 1 45 | + | M SBL 1 1 45 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 45 -0.0907 0.9581 | + | M SBV 1 45 -0.0907 0.9581 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FAAGA0004 | + | ID FL5FAAGA0004 |
| − | FORMULA C26H28O15 | + | FORMULA C26H28O15 |
| − | EXACTMASS 580.1428202259999 | + | EXACTMASS 580.1428202259999 |
| − | AVERAGEMASS 580.49152 | + | AVERAGEMASS 580.49152 |
| − | SMILES c(O4)(c(C(C(=C4c(c5)ccc(c5)O)OC(O2)C(OC(C3O)OCC(C3O)O)C(O)C(C2CO)O)=O)1)cc(O)cc1O | + | SMILES c(O4)(c(C(C(=C4c(c5)ccc(c5)O)OC(O2)C(OC(C3O)OCC(C3O)O)C(O)C(C2CO)O)=O)1)cc(O)cc1O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
41 45 0 0 0 0 0 0 0 0999 V2000
-3.5706 1.9717 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5706 1.1621 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8695 0.7574 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1685 1.1621 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1685 1.9717 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8695 2.3764 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4674 0.7574 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7664 1.1621 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7664 1.9717 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4674 2.3764 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4674 0.1262 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0655 2.3763 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6490 1.9637 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3634 2.3763 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3634 3.2014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6490 3.6138 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0655 3.2014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8695 -0.0519 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2714 2.3763 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0777 3.6137 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2114 -2.1359 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5803 -2.5930 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3290 -1.7140 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5803 -0.8297 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2114 -0.3725 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0397 -1.2516 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2114 0.4440 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4809 0.0126 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7889 -2.0721 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2413 0.4283 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8910 -0.4294 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8267 -0.0656 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7297 -0.0559 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0736 0.6004 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2550 0.1683 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1063 -0.3977 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5513 -0.4528 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.2714 0.2311 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3769 -2.3824 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1206 -3.0940 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7798 -3.6138 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
1 19 1 0 0 0 0
15 20 1 0 0 0 0
22 21 1 1 0 0 0
22 23 1 0 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 26 1 1 0 0 0
26 21 1 1 0 0 0
25 27 1 0 0 0 0
24 28 1 0 0 0 0
23 29 1 0 0 0 0
30 31 1 1 0 0 0
31 32 1 1 0 0 0
33 32 1 1 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 30 1 0 0 0 0
31 36 1 0 0 0 0
32 37 1 0 0 0 0
30 28 1 0 0 0 0
33 38 1 0 0 0 0
22 39 1 0 0 0 0
27 8 1 0 0 0 0
40 41 1 0 0 0 0
21 40 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 40 41
M SBL 1 1 45
M SMT 1 CH2OH
M SBV 1 45 -0.0907 0.9581
S SKP 5
ID FL5FAAGA0004
FORMULA C26H28O15
EXACTMASS 580.1428202259999
AVERAGEMASS 580.49152
SMILES c(O4)(c(C(C(=C4c(c5)ccc(c5)O)OC(O2)C(OC(C3O)OCC(C3O)O)C(O)C(C2CO)O)=O)1)cc(O)cc1O
M END