Mol:FL4DA9GS0001
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 42 46 0 0 0 0 0 0 0 0999 V2000 | + | 42 46 0 0 0 0 0 0 0 0999 V2000 |
| − | 3.8692 0.7405 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8692 0.7405 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4403 1.0702 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4403 1.0702 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4403 1.7296 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4403 1.7296 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8692 2.0593 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8692 2.0593 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2982 1.7296 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2982 1.7296 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2982 1.0702 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2982 1.0702 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3942 0.0475 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3942 0.0475 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9827 -0.2922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9827 -0.2922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5712 0.0475 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.5712 0.0475 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.5712 0.7270 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 2.5712 0.7270 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 1.9827 1.0668 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9827 1.0668 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3942 0.7270 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3942 0.7270 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8058 1.0668 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8058 1.0668 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2173 0.7270 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2173 0.7270 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2173 0.0475 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2173 0.0475 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8058 -0.2922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8058 -0.2922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2381 0.9900 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2381 0.9900 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8058 -0.8448 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8058 -0.8448 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9827 -0.8769 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9827 -0.8769 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0231 -0.2134 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0231 -0.2134 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5175 -1.2641 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -1.5175 -1.2641 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -1.0109 -1.7038 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.0109 -1.7038 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -0.4531 -1.3314 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -0.4531 -1.3314 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 0.2167 -1.2953 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 0.2167 -1.2953 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -0.2898 -0.8556 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2898 -0.8556 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8477 -1.2279 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -0.8477 -1.2279 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -0.0522 -1.7323 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0522 -1.7323 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8980 -2.3448 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8980 -2.3448 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0516 -1.5725 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0516 -1.5725 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2039 -1.2258 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -4.2039 -1.2258 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -3.8879 -1.7731 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -3.8879 -1.7731 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -3.2766 -1.6130 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -3.2766 -1.6130 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.6690 -1.7867 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.6690 -1.7867 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.9849 -1.2393 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9849 -1.2393 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5963 -1.3994 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -3.5963 -1.3994 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -4.4403 -1.6251 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4403 -1.6251 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3215 -2.1264 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3215 -2.1264 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.1432 -0.8826 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.1432 -0.8826 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2462 -0.4540 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2462 -0.4540 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2450 -0.4059 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2450 -0.4059 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5485 -0.9872 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5485 -0.9872 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3512 -0.3908 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3512 -0.3908 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 11 1 0 0 0 0 | + | 10 11 1 0 0 0 0 |
| − | 11 12 1 0 0 0 0 | + | 11 12 1 0 0 0 0 |
| − | 12 7 2 0 0 0 0 | + | 12 7 2 0 0 0 0 |
| − | 10 6 1 6 0 0 0 | + | 10 6 1 6 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 7 1 0 0 0 0 | + | 16 7 1 0 0 0 0 |
| − | 14 17 1 0 0 0 0 | + | 14 17 1 0 0 0 0 |
| − | 16 18 1 0 0 0 0 | + | 16 18 1 0 0 0 0 |
| − | 8 19 2 0 0 0 0 | + | 8 19 2 0 0 0 0 |
| − | 9 20 1 1 0 0 0 | + | 9 20 1 1 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 18 24 1 0 0 0 0 | + | 18 24 1 0 0 0 0 |
| − | 23 27 1 0 0 0 0 | + | 23 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 21 29 1 0 0 0 0 | + | 21 29 1 0 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 33 32 1 1 0 0 0 | + | 33 32 1 1 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 30 1 0 0 0 0 | + | 35 30 1 0 0 0 0 |
| − | 29 33 1 0 0 0 0 | + | 29 33 1 0 0 0 0 |
| − | 31 36 1 0 0 0 0 | + | 31 36 1 0 0 0 0 |
| − | 32 37 1 0 0 0 0 | + | 32 37 1 0 0 0 0 |
| − | 30 38 1 0 0 0 0 | + | 30 38 1 0 0 0 0 |
| − | 26 39 1 0 0 0 0 | + | 26 39 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 35 41 1 0 0 0 0 | + | 35 41 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 41 42 | + | M SAL 2 2 41 42 |
| − | M SBL 2 1 45 | + | M SBL 2 1 45 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SVB 2 45 -3.3788 -1.046 | + | M SVB 2 45 -3.3788 -1.046 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 39 40 | + | M SAL 1 2 39 40 |
| − | M SBL 1 1 43 | + | M SBL 1 1 43 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SVB 1 43 -1.2462 -0.454 | + | M SVB 1 43 -1.2462 -0.454 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL4DA9GS0001 | + | ID FL4DA9GS0001 |
| − | KNApSAcK_ID C00008666 | + | KNApSAcK_ID C00008666 |
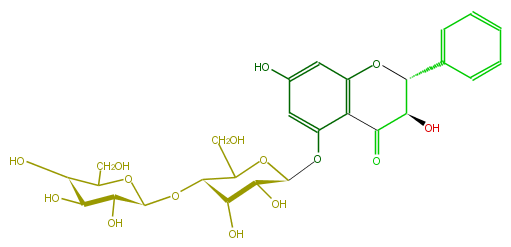
| − | NAME Pinobanksin 5-galactosyl-(1->4)-glucoside | + | NAME Pinobanksin 5-galactosyl-(1->4)-glucoside |
| − | CAS_RN 91925-95-0 | + | CAS_RN 91925-95-0 |
| − | FORMULA C27H32O15 | + | FORMULA C27H32O15 |
| − | EXACTMASS 596.174120354 | + | EXACTMASS 596.174120354 |
| − | AVERAGEMASS 596.5339799999999 | + | AVERAGEMASS 596.5339799999999 |
| − | SMILES [C@@H]([C@@H]1O[C@@H](C5CO)[C@@H]([C@H](O)[C@@H](O5)Oc(c4)c(C3=O)c(cc4O)O[C@@H]([C@@H](O)3)c(c2)cccc2)O)(O)[C@@H](O)[C@@H](O)C(O1)CO | + | SMILES [C@@H]([C@@H]1O[C@@H](C5CO)[C@@H]([C@H](O)[C@@H](O5)Oc(c4)c(C3=O)c(cc4O)O[C@@H]([C@@H](O)3)c(c2)cccc2)O)(O)[C@@H](O)[C@@H](O)C(O1)CO |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
42 46 0 0 0 0 0 0 0 0999 V2000
3.8692 0.7405 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4403 1.0702 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4403 1.7296 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8692 2.0593 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2982 1.7296 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2982 1.0702 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3942 0.0475 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9827 -0.2922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5712 0.0475 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.5712 0.7270 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
1.9827 1.0668 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3942 0.7270 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8058 1.0668 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2173 0.7270 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2173 0.0475 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8058 -0.2922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2381 0.9900 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8058 -0.8448 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9827 -0.8769 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0231 -0.2134 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.5175 -1.2641 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-1.0109 -1.7038 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.4531 -1.3314 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
0.2167 -1.2953 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.2898 -0.8556 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8477 -1.2279 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-0.0522 -1.7323 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8980 -2.3448 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0516 -1.5725 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2039 -1.2258 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-3.8879 -1.7731 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-3.2766 -1.6130 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.6690 -1.7867 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.9849 -1.2393 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5963 -1.3994 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-4.4403 -1.6251 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3215 -2.1264 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.1432 -0.8826 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2462 -0.4540 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2450 -0.4059 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5485 -0.9872 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3512 -0.3908 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 1 0 0 0 0
10 11 1 0 0 0 0
11 12 1 0 0 0 0
12 7 2 0 0 0 0
10 6 1 6 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 7 1 0 0 0 0
14 17 1 0 0 0 0
16 18 1 0 0 0 0
8 19 2 0 0 0 0
9 20 1 1 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
18 24 1 0 0 0 0
23 27 1 0 0 0 0
22 28 1 0 0 0 0
21 29 1 0 0 0 0
30 31 1 1 0 0 0
31 32 1 1 0 0 0
33 32 1 1 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 30 1 0 0 0 0
29 33 1 0 0 0 0
31 36 1 0 0 0 0
32 37 1 0 0 0 0
30 38 1 0 0 0 0
26 39 1 0 0 0 0
39 40 1 0 0 0 0
35 41 1 0 0 0 0
41 42 1 0 0 0 0
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 41 42
M SBL 2 1 45
M SMT 2 CH2OH
M SVB 2 45 -3.3788 -1.046
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 39 40
M SBL 1 1 43
M SMT 1 CH2OH
M SVB 1 43 -1.2462 -0.454
S SKP 8
ID FL4DA9GS0001
KNApSAcK_ID C00008666
NAME Pinobanksin 5-galactosyl-(1->4)-glucoside
CAS_RN 91925-95-0
FORMULA C27H32O15
EXACTMASS 596.174120354
AVERAGEMASS 596.5339799999999
SMILES [C@@H]([C@@H]1O[C@@H](C5CO)[C@@H]([C@H](O)[C@@H](O5)Oc(c4)c(C3=O)c(cc4O)O[C@@H]([C@@H](O)3)c(c2)cccc2)O)(O)[C@@H](O)[C@@H](O)C(O1)CO
M END