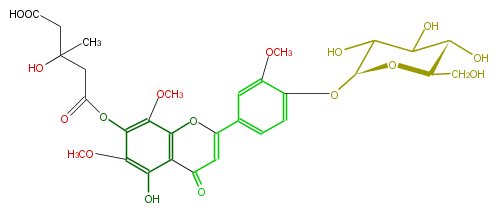
Mol:FL3FGCGS0007
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 47 50 0 0 0 0 0 0 0 0999 V2000 | + | 47 50 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.0546 2.8714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0546 2.8714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5116 2.7259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5116 2.7259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1434 3.0942 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1434 3.0942 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2782 3.5973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2782 3.5973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7812 3.7321 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7812 3.7321 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1496 3.3638 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1496 3.3638 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9099 3.9656 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9099 3.9656 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0447 4.4687 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0447 4.4687 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5477 4.6035 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5477 4.6035 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9161 4.2352 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9161 4.2352 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5101 4.0362 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5101 4.0362 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6826 5.1064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6826 5.1064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3072 5.4817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3072 5.4817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4446 5.9946 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4446 5.9946 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9573 6.1319 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9573 6.1319 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3326 5.7565 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3326 5.7565 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1953 5.2438 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1953 5.2438 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6413 2.9596 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6413 2.9596 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4605 2.5113 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4605 2.5113 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6985 7.0979 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6985 7.0979 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5100 8.1437 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -2.5100 8.1437 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -1.9618 7.5955 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.9618 7.5955 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.9746 8.3707 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9746 8.3707 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5748 9.0400 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.5748 9.0400 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.1230 9.5883 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.1230 9.5883 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.1102 8.8131 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -2.1102 8.8131 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -2.5100 7.3984 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5100 7.3984 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5443 9.2471 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5443 9.2471 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6663 10.0451 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6663 10.0451 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9028 2.2560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9028 2.2560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6169 1.7608 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6169 1.7608 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4539 2.4037 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4539 2.4037 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8574 2.0002 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -3.8574 2.0002 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -4.4085 2.1479 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4085 2.1479 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8120 1.7444 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8120 1.7444 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.3633 1.8921 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.3633 1.8921 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7162 1.3865 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7162 1.3865 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7355 1.5452 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7355 1.5452 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0100 2.5697 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0100 2.5697 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4680 9.5417 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4680 9.5417 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8847 10.1250 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8847 10.1250 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6193 3.7369 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6193 3.7369 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4024 4.3588 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4024 4.3588 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8505 6.0694 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8505 6.0694 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7064 6.5865 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7064 6.5865 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8241 2.0699 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8241 2.0699 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2541 1.1671 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2541 1.1671 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 20 15 1 0 0 0 0 | + | 20 15 1 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 26 28 1 0 0 0 0 | + | 26 28 1 0 0 0 0 |
| − | 25 29 1 0 0 0 0 | + | 25 29 1 0 0 0 0 |
| − | 22 20 1 0 0 0 0 | + | 22 20 1 0 0 0 0 |
| − | 19 30 1 0 0 0 0 | + | 19 30 1 0 0 0 0 |
| − | 30 31 2 0 0 0 0 | + | 30 31 2 0 0 0 0 |
| − | 30 32 1 0 0 0 0 | + | 30 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 35 37 2 0 0 0 0 | + | 35 37 2 0 0 0 0 |
| − | 33 38 1 0 0 0 0 | + | 33 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 24 40 1 0 0 0 0 | + | 24 40 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 6 42 1 0 0 0 0 | + | 6 42 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 16 44 1 0 0 0 0 | + | 16 44 1 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 2 46 1 0 0 0 0 | + | 2 46 1 0 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | M STY 1 5 SUP | + | M STY 1 5 SUP |
| − | M SLB 1 5 5 | + | M SLB 1 5 5 |
| − | M SAL 5 2 40 41 | + | M SAL 5 2 40 41 |
| − | M SBL 5 1 43 | + | M SBL 5 1 43 |
| − | M SMT 5 CH2OH | + | M SMT 5 CH2OH |
| − | M SVB 5 43 4.0018 0.3597 | + | M SVB 5 43 4.0018 0.3597 |
| − | M STY 1 4 SUP | + | M STY 1 4 SUP |
| − | M SLB 1 4 4 | + | M SLB 1 4 4 |
| − | M SAL 4 2 46 47 | + | M SAL 4 2 46 47 |
| − | M SBL 4 1 49 | + | M SBL 4 1 49 |
| − | M SMT 4 OCH3 | + | M SMT 4 OCH3 |
| − | M SVB 4 49 -3.3075 -1.2302 | + | M SVB 4 49 -3.3075 -1.2302 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 44 45 | + | M SAL 3 2 44 45 |
| − | M SBL 3 1 47 | + | M SBL 3 1 47 |
| − | M SMT 3 OCH3 | + | M SMT 3 OCH3 |
| − | M SVB 3 47 0.29 0.7964 | + | M SVB 3 47 0.29 0.7964 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 42 43 | + | M SAL 2 2 42 43 |
| − | M SBL 2 1 45 | + | M SBL 2 1 45 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SVB 2 45 -1.9032 -0.0306 | + | M SVB 2 45 -1.9032 -0.0306 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 3 35 37 36 | + | M SAL 1 3 35 37 36 |
| − | M SBL 1 1 38 | + | M SBL 1 1 38 |
| − | M SMT 1 COOH | + | M SMT 1 COOH |
| − | M SVB 1 38 -4.3953 1.5717 | + | M SVB 1 38 -4.3953 1.5717 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL3FGCGS0007 | + | ID FL3FGCGS0007 |
| − | KNApSAcK_ID C00004445 | + | KNApSAcK_ID C00004445 |
| − | NAME 5,7,4'-Trihydroxy-6,8,3'-trimethoxyflavone 7-(3-hydroxy-3-methylglutarate)-4'-glucoside | + | NAME 5,7,4'-Trihydroxy-6,8,3'-trimethoxyflavone 7-(3-hydroxy-3-methylglutarate)-4'-glucoside |
| − | CAS_RN 101899-84-7 | + | CAS_RN 101899-84-7 |
| − | FORMULA C30H34O17 | + | FORMULA C30H34O17 |
| − | EXACTMASS 666.179599662 | + | EXACTMASS 666.179599662 |
| − | AVERAGEMASS 666.58076 | + | AVERAGEMASS 666.58076 |
| − | SMILES C(O)(=O)CC(CC(Oc(c(OC)4)c(OC)c(O)c(c41)C(=O)C=C(c(c2)ccc(O[C@H](O3)C(O)C([C@H]([C@H]3CO)O)O)c(OC)2)O1)=O)(C)O | + | SMILES C(O)(=O)CC(CC(Oc(c(OC)4)c(OC)c(O)c(c41)C(=O)C=C(c(c2)ccc(O[C@H](O3)C(O)C([C@H]([C@H]3CO)O)O)c(OC)2)O1)=O)(C)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/ 47 50 0 0 0 0 0 0 0 0999 V2000 -2.0546 2.8714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -1.5116 2.7259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -1.1434 3.0942 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -1.2782 3.5973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -1.7812 3.7321 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -2.1496 3.3638 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -0.9099 3.9656 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -1.0447 4.4687 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -1.5477 4.6035 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -1.9161 4.2352 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 -0.5101 4.0362 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 -1.6826 5.1064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -1.3072 5.4817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -1.4446 5.9946 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -1.9573 6.1319 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -2.3326 5.7565 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -2.1953 5.2438 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -0.6413 2.9596 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 -2.4605 2.5113 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 -1.6985 7.0979 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 -2.5100 8.1437 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 -1.9618 7.5955 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 -1.9746 8.3707 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 -1.5748 9.0400 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 -2.1230 9.5883 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 -2.1102 8.8131 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 -2.5100 7.3984 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 -2.5443 9.2471 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 -1.6663 10.0451 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 -2.9028 2.2560 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -2.6169 1.7608 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 -3.4539 2.4037 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -3.8574 2.0002 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 -4.4085 2.1479 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -4.8120 1.7444 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -5.3633 1.8921 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 -4.7162 1.3865 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 -3.7355 1.5452 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 -4.0100 2.5697 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -1.4680 9.5417 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -0.8847 10.1250 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 -2.6193 3.7369 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 -3.4024 4.3588 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -2.8505 6.0694 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 -3.7064 6.5865 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 -1.8241 2.0699 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 -2.2541 1.1671 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 1 2 1 0 0 0 0 2 3 2 0 0 0 0 3 4 1 0 0 0 0 4 5 2 0 0 0 0 5 6 1 0 0 0 0 6 1 2 0 0 0 0 4 7 1 0 0 0 0 7 8 1 0 0 0 0 8 9 2 0 0 0 0 9 10 1 0 0 0 0 10 5 1 0 0 0 0 7 11 2 0 0 0 0 9 12 1 0 0 0 0 12 13 2 0 0 0 0 13 14 1 0 0 0 0 14 15 2 0 0 0 0 15 16 1 0 0 0 0 16 17 2 0 0 0 0 17 12 1 0 0 0 0 3 18 1 0 0 0 0 1 19 1 0 0 0 0 20 15 1 0 0 0 0 21 22 1 0 0 0 0 22 23 1 1 0 0 0 23 24 1 1 0 0 0 25 24 1 1 0 0 0 25 26 1 0 0 0 0 26 21 1 0 0 0 0 21 27 1 0 0 0 0 26 28 1 0 0 0 0 25 29 1 0 0 0 0 22 20 1 0 0 0 0 19 30 1 0 0 0 0 30 31 2 0 0 0 0 30 32 1 0 0 0 0 32 33 1 0 0 0 0 33 34 1 0 0 0 0 34 35 1 0 0 0 0 35 36 1 0 0 0 0 35 37 2 0 0 0 0 33 38 1 0 0 0 0 33 39 1 0 0 0 0 24 40 1 0 0 0 0 40 41 1 0 0 0 0 6 42 1 0 0 0 0 42 43 1 0 0 0 0 16 44 1 0 0 0 0 44 45 1 0 0 0 0 2 46 1 0 0 0 0 46 47 1 0 0 0 0 M STY 1 5 SUP M SLB 1 5 5 M SAL 5 2 40 41 M SBL 5 1 43 M SMT 5 CH2OH M SVB 5 43 4.0018 0.3597 M STY 1 4 SUP M SLB 1 4 4 M SAL 4 2 46 47 M SBL 4 1 49 M SMT 4 OCH3 M SVB 4 49 -3.3075 -1.2302 M STY 1 3 SUP M SLB 1 3 3 M SAL 3 2 44 45 M SBL 3 1 47 M SMT 3 OCH3 M SVB 3 47 0.29 0.7964 M STY 1 2 SUP M SLB 1 2 2 M SAL 2 2 42 43 M SBL 2 1 45 M SMT 2 OCH3 M SVB 2 45 -1.9032 -0.0306 M STY 1 1 SUP M SLB 1 1 1 M SAL 1 3 35 37 36 M SBL 1 1 38 M SMT 1 COOH M SVB 1 38 -4.3953 1.5717 S SKP 8 ID FL3FGCGS0007 KNApSAcK_ID C00004445 NAME 5,7,4'-Trihydroxy-6,8,3'-trimethoxyflavone 7-(3-hydroxy-3-methylglutarate)-4'-glucoside CAS_RN 101899-84-7 FORMULA C30H34O17 EXACTMASS 666.179599662 AVERAGEMASS 666.58076 SMILES C(O)(=O)CC(CC(Oc(c(OC)4)c(OC)c(O)c(c41)C(=O)C=C(c(c2)ccc(O[C@H](O3)C(O)C([C@H]([C@H]3CO)O)O)c(OC)2)O1)=O)(C)O M END