Mol:FL3FECGS0034
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 42 46 0 0 0 0 0 0 0 0999 V2000 | + | 42 46 0 0 0 0 0 0 0 0999 V2000 |
| − | 0.6598 -0.3447 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6598 -0.3447 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6598 -1.1714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6598 -1.1714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3232 -1.5544 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3232 -1.5544 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9865 -1.1714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9865 -1.1714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9865 -0.4055 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9865 -0.4055 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3232 -0.0223 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3232 -0.0223 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6499 -1.5544 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6499 -1.5544 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3133 -1.1714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3133 -1.1714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3133 -0.4055 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3133 -0.4055 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6499 -0.0223 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6499 -0.0223 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6499 -2.1516 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6499 -2.1516 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9764 -0.0225 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9764 -0.0225 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6525 -0.4130 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6525 -0.4130 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.3286 -0.0225 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.3286 -0.0225 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.3286 0.7581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.3286 0.7581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6525 1.1485 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6525 1.1485 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9764 0.7581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9764 0.7581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3232 -2.3188 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3232 -2.3188 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0185 0.0469 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0185 0.0469 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.0047 1.1485 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.0047 1.1485 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3181 1.9486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3181 1.9486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1618 0.7025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1618 0.7025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0038 0.5242 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0038 0.5242 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0856 0.0074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0856 0.0074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3658 1.0534 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3658 1.0534 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5321 1.2643 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5321 1.2643 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2338 1.9679 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2338 1.9679 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9860 1.3249 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9860 1.3249 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5938 -0.3901 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5938 -0.3901 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.0047 1.1761 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.0047 1.1761 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.3139 1.2090 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.3139 1.2090 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.1575 -0.0371 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.1575 -0.0371 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9995 -0.2154 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9995 -0.2154 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0814 -0.7324 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0814 -0.7324 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3615 0.3138 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3615 0.3138 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5279 0.5246 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5279 0.5246 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.8104 0.3383 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.8104 0.3383 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6071 -1.0372 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6071 -1.0372 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6525 1.9291 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6525 1.9291 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0547 -1.5839 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0547 -1.5839 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9073 2.3188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9073 2.3188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3006 1.2680 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3006 1.2680 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 1 1 0 0 0 0 | + | 19 1 1 0 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 32 37 1 0 0 0 0 | + | 32 37 1 0 0 0 0 |
| − | 33 38 1 0 0 0 0 | + | 33 38 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 34 29 1 0 0 0 0 | + | 34 29 1 0 0 0 0 |
| − | 24 19 1 0 0 0 0 | + | 24 19 1 0 0 0 0 |
| − | 16 39 1 0 0 0 0 | + | 16 39 1 0 0 0 0 |
| − | 2 40 1 0 0 0 0 | + | 2 40 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 26 41 1 0 0 0 0 | + | 26 41 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 41 42 | + | M SAL 1 2 41 42 |
| − | M SBL 1 1 46 | + | M SBL 1 1 46 |
| − | M SMT 1 ^CH2OH | + | M SMT 1 ^CH2OH |
| − | M SBV 1 46 0.3753 -1.0545 | + | M SBV 1 46 0.3753 -1.0545 |
| − | S SKP 5 | + | S SKP 5 |
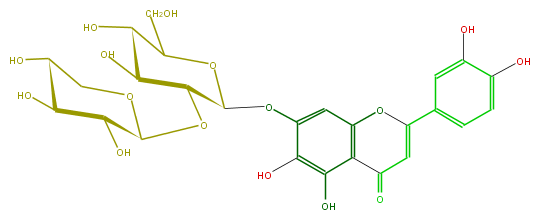
| − | ID FL3FECGS0034 | + | ID FL3FECGS0034 |
| − | FORMULA C26H28O16 | + | FORMULA C26H28O16 |
| − | EXACTMASS 596.137734848 | + | EXACTMASS 596.137734848 |
| − | AVERAGEMASS 596.49092 | + | AVERAGEMASS 596.49092 |
| − | SMILES C(C5O)(O)C(OCC5O)OC(C(O)4)C(OC(CO)C4O)Oc(c1)c(O)c(O)c(C2=O)c1OC(c(c3)ccc(c3O)O)=C2 | + | SMILES C(C5O)(O)C(OCC5O)OC(C(O)4)C(OC(CO)C4O)Oc(c1)c(O)c(O)c(C2=O)c1OC(c(c3)ccc(c3O)O)=C2 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
42 46 0 0 0 0 0 0 0 0999 V2000
0.6598 -0.3447 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6598 -1.1714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3232 -1.5544 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9865 -1.1714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9865 -0.4055 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3232 -0.0223 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6499 -1.5544 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3133 -1.1714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3133 -0.4055 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6499 -0.0223 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6499 -2.1516 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.9764 -0.0225 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.6525 -0.4130 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.3286 -0.0225 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.3286 0.7581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.6525 1.1485 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9764 0.7581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3232 -2.3188 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0185 0.0469 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
6.0047 1.1485 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3181 1.9486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1618 0.7025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0038 0.5242 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0856 0.0074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3658 1.0534 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5321 1.2643 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2338 1.9679 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9860 1.3249 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.5938 -0.3901 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-6.0047 1.1761 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.3139 1.2090 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.1575 -0.0371 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9995 -0.2154 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0814 -0.7324 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3615 0.3138 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.5279 0.5246 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.8104 0.3383 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6071 -1.0372 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.6525 1.9291 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0547 -1.5839 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9073 2.3188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3006 1.2680 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 1 1 0 0 0 0
15 20 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
32 37 1 0 0 0 0
33 38 1 0 0 0 0
30 31 1 0 0 0 0
34 29 1 0 0 0 0
24 19 1 0 0 0 0
16 39 1 0 0 0 0
2 40 1 0 0 0 0
41 42 1 0 0 0 0
26 41 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 41 42
M SBL 1 1 46
M SMT 1 ^CH2OH
M SBV 1 46 0.3753 -1.0545
S SKP 5
ID FL3FECGS0034
FORMULA C26H28O16
EXACTMASS 596.137734848
AVERAGEMASS 596.49092
SMILES C(C5O)(O)C(OCC5O)OC(C(O)4)C(OC(CO)C4O)Oc(c1)c(O)c(O)c(C2=O)c1OC(c(c3)ccc(c3O)O)=C2
M END