Mol:FL3FEAGS0050
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 44 48 0 0 0 0 0 0 0 0999 V2000 | + | 44 48 0 0 0 0 0 0 0 0999 V2000 |
| − | 1.2892 -0.3519 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2892 -0.3519 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2892 -1.1769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2892 -1.1769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0037 -1.5894 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0037 -1.5894 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7181 -1.1769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7181 -1.1769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7181 -0.3519 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7181 -0.3519 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0036 0.0606 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0036 0.0606 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4326 -1.5894 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4326 -1.5894 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1471 -1.1769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1471 -1.1769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1471 -0.3519 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1471 -0.3519 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4326 0.0606 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4326 0.0606 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8615 0.0606 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8615 0.0606 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.5760 -0.3519 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.5760 -0.3519 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.2905 0.0606 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.2905 0.0606 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.2905 0.8856 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.2905 0.8856 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.5760 1.2981 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.5760 1.2981 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8615 0.8856 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8615 0.8856 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4326 -2.4144 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4326 -2.4144 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0037 -2.4144 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0037 -2.4144 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 7.0368 1.3165 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 7.0368 1.3165 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5696 -0.0049 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5696 -0.0049 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6022 -1.5735 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6022 -1.5735 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1080 -1.1634 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1080 -1.1634 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2317 0.3854 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2317 0.3854 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8190 -0.3293 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8190 -0.3293 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0207 -0.1201 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0207 -0.1201 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2272 -0.3470 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2272 -0.3470 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6398 0.3678 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6398 0.3678 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4382 0.1587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4382 0.1587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5952 0.7445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5952 0.7445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7149 -0.0978 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7149 -0.0978 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2677 -0.0702 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2677 -0.0702 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1922 -0.7601 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1922 -0.7601 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0912 0.9874 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0912 0.9874 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5032 1.7009 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5032 1.7009 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0912 2.4144 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0912 2.4144 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3271 1.7009 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3271 1.7009 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7390 0.9874 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7390 0.9874 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5629 0.9874 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5629 0.9874 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9754 1.7018 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9754 1.7018 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.8004 1.7018 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.8004 1.7018 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.2129 0.9874 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.2129 0.9874 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.8004 0.2729 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.8004 0.2729 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9754 0.2729 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9754 0.2729 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -7.0368 0.9874 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -7.0368 0.9874 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 9 11 1 0 0 0 0 | + | 9 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 7 17 2 0 0 0 0 | + | 7 17 2 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 14 19 1 0 0 0 0 | + | 14 19 1 0 0 0 0 |
| − | 20 1 1 0 0 0 0 | + | 20 1 1 0 0 0 0 |
| − | 2 21 1 0 0 0 0 | + | 2 21 1 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 23 1 0 0 0 0 | + | 28 23 1 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 23 30 1 0 0 0 0 | + | 23 30 1 0 0 0 0 |
| − | 24 31 1 0 0 0 0 | + | 24 31 1 0 0 0 0 |
| − | 25 32 1 0 0 0 0 | + | 25 32 1 0 0 0 0 |
| − | 26 20 1 0 0 0 0 | + | 26 20 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 2 0 0 0 0 | + | 34 35 2 0 0 0 0 |
| − | 34 36 1 0 0 0 0 | + | 34 36 1 0 0 0 0 |
| − | 36 37 2 0 0 0 0 | + | 36 37 2 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 2 0 0 0 0 | + | 38 39 2 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 40 41 2 0 0 0 0 | + | 40 41 2 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 42 43 2 0 0 0 0 | + | 42 43 2 0 0 0 0 |
| − | 43 38 1 0 0 0 0 | + | 43 38 1 0 0 0 0 |
| − | 41 44 1 0 0 0 0 | + | 41 44 1 0 0 0 0 |
| − | 33 29 1 0 0 0 0 | + | 33 29 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL3FEAGS0050 | + | ID FL3FEAGS0050 |
| − | KNApSAcK_ID C00013637 | + | KNApSAcK_ID C00013637 |
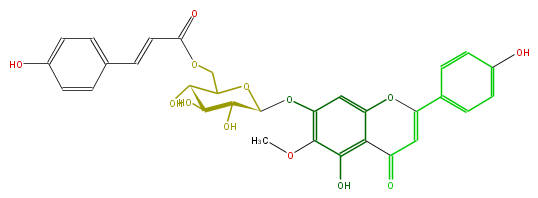
| − | NAME Hispidulin 7-(6''-E-p-Coumaroylglucoside);5-Hydroxy-2-(4-hydroxyphenyl)-7-[[6-O-[(2E)-3-(4-hydroxyphenyl)-1-oxo-2-propenyl]-beta-D-glucopyranosyl]oxy]-6-methoxy-4H-1-benzopyran-4-one | + | NAME Hispidulin 7-(6''-E-p-Coumaroylglucoside);5-Hydroxy-2-(4-hydroxyphenyl)-7-[[6-O-[(2E)-3-(4-hydroxyphenyl)-1-oxo-2-propenyl]-beta-D-glucopyranosyl]oxy]-6-methoxy-4H-1-benzopyran-4-one |
| − | CAS_RN 487040-42-6 | + | CAS_RN 487040-42-6 |
| − | FORMULA C31H28O13 | + | FORMULA C31H28O13 |
| − | EXACTMASS 608.152990982 | + | EXACTMASS 608.152990982 |
| − | AVERAGEMASS 608.5462200000001 | + | AVERAGEMASS 608.5462200000001 |
| − | SMILES C(O)(C5O)C(OC(C5O)COC(C=Cc(c4)ccc(O)c4)=O)Oc(c(OC)1)cc(O2)c(C(=O)C=C(c(c3)ccc(c3)O)2)c1O | + | SMILES C(O)(C5O)C(OC(C5O)COC(C=Cc(c4)ccc(O)c4)=O)Oc(c(OC)1)cc(O2)c(C(=O)C=C(c(c3)ccc(c3)O)2)c1O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
44 48 0 0 0 0 0 0 0 0999 V2000
1.2892 -0.3519 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2892 -1.1769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0037 -1.5894 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7181 -1.1769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7181 -0.3519 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0036 0.0606 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4326 -1.5894 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1471 -1.1769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1471 -0.3519 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4326 0.0606 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.8615 0.0606 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.5760 -0.3519 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.2905 0.0606 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.2905 0.8856 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.5760 1.2981 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.8615 0.8856 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4326 -2.4144 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0037 -2.4144 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
7.0368 1.3165 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5696 -0.0049 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6022 -1.5735 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1080 -1.1634 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2317 0.3854 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8190 -0.3293 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0207 -0.1201 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2272 -0.3470 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6398 0.3678 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4382 0.1587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5952 0.7445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7149 -0.0978 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2677 -0.0702 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.1922 -0.7601 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0912 0.9874 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5032 1.7009 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0912 2.4144 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3271 1.7009 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7390 0.9874 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.5629 0.9874 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.9754 1.7018 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.8004 1.7018 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.2129 0.9874 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.8004 0.2729 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.9754 0.2729 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-7.0368 0.9874 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
7 17 2 0 0 0 0
3 18 1 0 0 0 0
14 19 1 0 0 0 0
20 1 1 0 0 0 0
2 21 1 0 0 0 0
21 22 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 28 1 0 0 0 0
28 23 1 0 0 0 0
28 29 1 0 0 0 0
23 30 1 0 0 0 0
24 31 1 0 0 0 0
25 32 1 0 0 0 0
26 20 1 0 0 0 0
33 34 1 0 0 0 0
34 35 2 0 0 0 0
34 36 1 0 0 0 0
36 37 2 0 0 0 0
37 38 1 0 0 0 0
38 39 2 0 0 0 0
39 40 1 0 0 0 0
40 41 2 0 0 0 0
41 42 1 0 0 0 0
42 43 2 0 0 0 0
43 38 1 0 0 0 0
41 44 1 0 0 0 0
33 29 1 0 0 0 0
S SKP 8
ID FL3FEAGS0050
KNApSAcK_ID C00013637
NAME Hispidulin 7-(6''-E-p-Coumaroylglucoside);5-Hydroxy-2-(4-hydroxyphenyl)-7-[[6-O-[(2E)-3-(4-hydroxyphenyl)-1-oxo-2-propenyl]-beta-D-glucopyranosyl]oxy]-6-methoxy-4H-1-benzopyran-4-one
CAS_RN 487040-42-6
FORMULA C31H28O13
EXACTMASS 608.152990982
AVERAGEMASS 608.5462200000001
SMILES C(O)(C5O)C(OC(C5O)COC(C=Cc(c4)ccc(O)c4)=O)Oc(c(OC)1)cc(O2)c(C(=O)C=C(c(c3)ccc(c3)O)2)c1O
M END