Mol:FL3FCACS0026
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 43 47 0 0 0 0 0 0 0 0999 V2000 | + | 43 47 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.0054 -0.7261 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0054 -0.7261 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0054 -1.3685 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0054 -1.3685 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4491 -1.6897 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4491 -1.6897 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1072 -1.3685 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1072 -1.3685 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1072 -0.7261 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1072 -0.7261 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4491 -0.4049 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4491 -0.4049 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6635 -1.6897 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6635 -1.6897 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2198 -1.3685 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2198 -1.3685 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2198 -0.7261 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2198 -0.7261 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6635 -0.4049 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6635 -0.4049 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6635 -2.1905 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6635 -2.1905 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2037 2.2795 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -0.2037 2.2795 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -0.8093 1.8208 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -0.8093 1.8208 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -0.5524 1.1601 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -0.5524 1.1601 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -0.5455 0.5226 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -0.5455 0.5226 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -0.0822 0.9859 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0822 0.9859 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3872 1.5638 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -0.3872 1.5638 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 0.2971 2.2795 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2971 2.2795 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8441 2.5359 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8441 2.5359 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1878 0.7816 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1878 0.7816 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4491 -2.3318 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4491 -2.3318 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8378 -0.3844 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8378 -0.3844 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4240 -0.7229 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4240 -0.7229 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0102 -0.3844 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0102 -0.3844 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0102 0.2925 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0102 0.2925 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4240 0.6309 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4240 0.6309 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8378 0.2925 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8378 0.2925 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5961 0.6307 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5961 0.6307 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0822 -1.7397 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -3.0822 -1.7397 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -2.7111 -2.2297 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.7111 -2.2297 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.1766 -2.0218 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.1766 -2.0218 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.6608 -2.0163 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.6608 -2.0163 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.0356 -1.6414 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0356 -1.6414 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5032 -1.8882 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -2.5032 -1.8882 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -3.6732 -1.6880 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6732 -1.6880 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0614 -2.6909 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0614 -2.6909 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8703 -2.5359 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8703 -2.5359 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0364 1.8084 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0364 1.8084 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7509 1.3959 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7509 1.3959 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3626 -0.1074 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3626 -0.1074 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8626 0.7586 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8626 0.7586 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1576 -1.0921 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1576 -1.0921 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5701 -0.3776 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5701 -0.3776 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 12 13 1 1 0 0 0 | + | 12 13 1 1 0 0 0 |
| − | 13 14 1 1 0 0 0 | + | 13 14 1 1 0 0 0 |
| − | 15 14 1 1 0 0 0 | + | 15 14 1 1 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 12 18 1 0 0 0 0 | + | 12 18 1 0 0 0 0 |
| − | 13 19 1 0 0 0 0 | + | 13 19 1 0 0 0 0 |
| − | 14 20 1 0 0 0 0 | + | 14 20 1 0 0 0 0 |
| − | 6 15 1 0 0 0 0 | + | 6 15 1 0 0 0 0 |
| − | 3 21 1 0 0 0 0 | + | 3 21 1 0 0 0 0 |
| − | 22 23 2 0 0 0 0 | + | 22 23 2 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 2 0 0 0 0 | + | 24 25 2 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 2 0 0 0 0 | + | 26 27 2 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 9 1 0 0 0 0 | + | 22 9 1 0 0 0 0 |
| − | 25 28 1 0 0 0 0 | + | 25 28 1 0 0 0 0 |
| − | 29 30 1 1 0 0 0 | + | 29 30 1 1 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 32 31 1 1 0 0 0 | + | 32 31 1 1 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 29 1 0 0 0 0 | + | 34 29 1 0 0 0 0 |
| − | 29 35 1 0 0 0 0 | + | 29 35 1 0 0 0 0 |
| − | 30 36 1 0 0 0 0 | + | 30 36 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 2 1 0 0 0 0 | + | 32 2 1 0 0 0 0 |
| − | 17 38 1 0 0 0 0 | + | 17 38 1 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 1 40 1 0 0 0 0 | + | 1 40 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 34 42 1 0 0 0 0 | + | 34 42 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 42 43 | + | M SAL 3 2 42 43 |
| − | M SBL 3 1 46 | + | M SBL 3 1 46 |
| − | M SMT 3 CH2OH | + | M SMT 3 CH2OH |
| − | M SVB 3 46 -2.7598 -0.8901 | + | M SVB 3 46 -2.7598 -0.8901 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 38 39 | + | M SAL 2 2 38 39 |
| − | M SBL 2 1 42 | + | M SBL 2 1 42 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SVB 2 42 0.0364 1.8084 | + | M SVB 2 42 0.0364 1.8084 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 40 41 | + | M SAL 1 2 40 41 |
| − | M SBL 1 1 44 | + | M SBL 1 1 44 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 44 -1.3626 -0.1074 | + | M SVB 1 44 -1.3626 -0.1074 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL3FCACS0026 | + | ID FL3FCACS0026 |
| − | KNApSAcK_ID C00006400 | + | KNApSAcK_ID C00006400 |
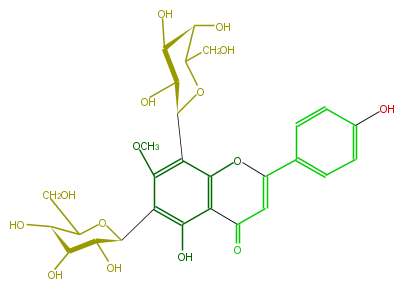
| − | NAME 6-C-Glucopyranosyl-8-C-galactopyranosylgenkwanin | + | NAME 6-C-Glucopyranosyl-8-C-galactopyranosylgenkwanin |
| − | CAS_RN - | + | CAS_RN - |
| − | FORMULA C28H32O15 | + | FORMULA C28H32O15 |
| − | EXACTMASS 608.174120354 | + | EXACTMASS 608.174120354 |
| − | AVERAGEMASS 608.54468 | + | AVERAGEMASS 608.54468 |
| − | SMILES c(C2=O)(c4O)c(c(c(c4[C@H](O5)[C@H]([C@H]([C@@H](O)C5CO)O)O)OC)[C@H](O3)[C@@H](O)[C@H]([C@H](C3CO)O)O)OC(=C2)c(c1)ccc(c1)O | + | SMILES c(C2=O)(c4O)c(c(c(c4[C@H](O5)[C@H]([C@H]([C@@H](O)C5CO)O)O)OC)[C@H](O3)[C@@H](O)[C@H]([C@H](C3CO)O)O)OC(=C2)c(c1)ccc(c1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
43 47 0 0 0 0 0 0 0 0999 V2000
-1.0054 -0.7261 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0054 -1.3685 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4491 -1.6897 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1072 -1.3685 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1072 -0.7261 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4491 -0.4049 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6635 -1.6897 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2198 -1.3685 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2198 -0.7261 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6635 -0.4049 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6635 -2.1905 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2037 2.2795 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-0.8093 1.8208 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.5524 1.1601 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.5455 0.5226 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.0822 0.9859 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.3872 1.5638 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
0.2971 2.2795 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8441 2.5359 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.1878 0.7816 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.4491 -2.3318 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8378 -0.3844 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4240 -0.7229 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0102 -0.3844 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0102 0.2925 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4240 0.6309 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8378 0.2925 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5961 0.6307 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0822 -1.7397 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-2.7111 -2.2297 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.1766 -2.0218 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.6608 -2.0163 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.0356 -1.6414 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5032 -1.8882 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-3.6732 -1.6880 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0614 -2.6909 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8703 -2.5359 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0364 1.8084 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7509 1.3959 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3626 -0.1074 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8626 0.7586 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1576 -1.0921 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5701 -0.3776 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
12 13 1 1 0 0 0
13 14 1 1 0 0 0
15 14 1 1 0 0 0
15 16 1 0 0 0 0
16 17 1 0 0 0 0
17 12 1 0 0 0 0
12 18 1 0 0 0 0
13 19 1 0 0 0 0
14 20 1 0 0 0 0
6 15 1 0 0 0 0
3 21 1 0 0 0 0
22 23 2 0 0 0 0
23 24 1 0 0 0 0
24 25 2 0 0 0 0
25 26 1 0 0 0 0
26 27 2 0 0 0 0
27 22 1 0 0 0 0
22 9 1 0 0 0 0
25 28 1 0 0 0 0
29 30 1 1 0 0 0
30 31 1 1 0 0 0
32 31 1 1 0 0 0
32 33 1 0 0 0 0
33 34 1 0 0 0 0
34 29 1 0 0 0 0
29 35 1 0 0 0 0
30 36 1 0 0 0 0
31 37 1 0 0 0 0
32 2 1 0 0 0 0
17 38 1 0 0 0 0
38 39 1 0 0 0 0
1 40 1 0 0 0 0
40 41 1 0 0 0 0
34 42 1 0 0 0 0
42 43 1 0 0 0 0
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 42 43
M SBL 3 1 46
M SMT 3 CH2OH
M SVB 3 46 -2.7598 -0.8901
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 38 39
M SBL 2 1 42
M SMT 2 CH2OH
M SVB 2 42 0.0364 1.8084
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 40 41
M SBL 1 1 44
M SMT 1 OCH3
M SVB 1 44 -1.3626 -0.1074
S SKP 8
ID FL3FCACS0026
KNApSAcK_ID C00006400
NAME 6-C-Glucopyranosyl-8-C-galactopyranosylgenkwanin
CAS_RN -
FORMULA C28H32O15
EXACTMASS 608.174120354
AVERAGEMASS 608.54468
SMILES c(C2=O)(c4O)c(c(c(c4[C@H](O5)[C@H]([C@H]([C@@H](O)C5CO)O)O)OC)[C@H](O3)[C@@H](O)[C@H]([C@H](C3CO)O)O)OC(=C2)c(c1)ccc(c1)O
M END