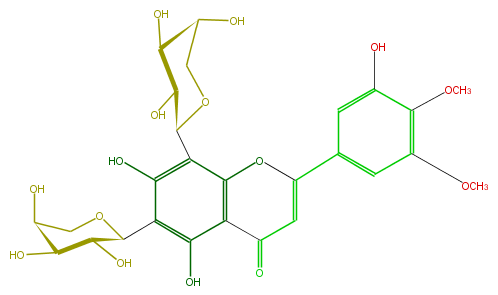
Mol:FL3FAKCS0001
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 42 46 0 0 0 0 0 0 0 0999 V2000 | + | 42 46 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.3637 -0.6728 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3637 -0.6728 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3637 -1.4977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3637 -1.4977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6493 -1.9102 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6493 -1.9102 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9348 -1.4977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9348 -1.4977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9348 -0.6728 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9348 -0.6728 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6493 -0.2602 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6493 -0.2602 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2203 -1.9102 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2203 -1.9102 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4941 -1.4977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4941 -1.4977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4941 -0.6728 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4941 -0.6728 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2203 -0.2602 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2203 -0.2602 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2203 -2.5535 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2203 -2.5535 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0779 -0.2604 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0779 -0.2604 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4844 2.5925 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4844 2.5925 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2621 2.0033 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2621 2.0033 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9321 1.1549 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9321 1.1549 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9234 0.3361 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9234 0.3361 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3282 0.9312 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3282 0.9312 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7201 1.6734 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7201 1.6734 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7544 2.5775 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7544 2.5775 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3055 2.7348 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3055 2.7348 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3551 0.6674 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3551 0.6674 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6493 -2.7348 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6493 -2.7348 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8308 -1.5266 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8308 -1.5266 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3541 -2.1559 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3541 -2.1559 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6678 -1.8889 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6678 -1.8889 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0053 -1.8817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0053 -1.8817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4867 -1.4004 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4867 -1.4004 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0872 -1.7173 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0872 -1.7173 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8317 -0.8394 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8317 -0.8394 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0973 -2.1920 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0973 -2.1920 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0668 -2.3443 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0668 -2.3443 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3672 -0.1279 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3672 -0.1279 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1201 -0.5626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1201 -0.5626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8729 -0.1279 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8729 -0.1279 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8729 0.7414 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8729 0.7414 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1201 1.1761 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1201 1.1761 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3672 0.7414 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3672 0.7414 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1201 2.0450 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1201 2.0450 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9851 -0.7701 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9851 -0.7701 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0973 -1.4124 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0973 -1.4124 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6254 1.1759 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6254 1.1759 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5430 0.6461 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5430 0.6461 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 1 12 1 0 0 0 0 | + | 1 12 1 0 0 0 0 |
| − | 13 14 1 1 0 0 0 | + | 13 14 1 1 0 0 0 |
| − | 14 15 1 1 0 0 0 | + | 14 15 1 1 0 0 0 |
| − | 16 15 1 1 0 0 0 | + | 16 15 1 1 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 18 1 0 0 0 0 | + | 17 18 1 0 0 0 0 |
| − | 18 13 1 0 0 0 0 | + | 18 13 1 0 0 0 0 |
| − | 13 19 1 0 0 0 0 | + | 13 19 1 0 0 0 0 |
| − | 14 20 1 0 0 0 0 | + | 14 20 1 0 0 0 0 |
| − | 15 21 1 0 0 0 0 | + | 15 21 1 0 0 0 0 |
| − | 6 16 1 0 0 0 0 | + | 6 16 1 0 0 0 0 |
| − | 3 22 1 0 0 0 0 | + | 3 22 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 23 1 0 0 0 0 | + | 28 23 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 25 31 1 0 0 0 0 | + | 25 31 1 0 0 0 0 |
| − | 26 2 1 0 0 0 0 | + | 26 2 1 0 0 0 0 |
| − | 32 33 2 0 0 0 0 | + | 32 33 2 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 2 0 0 0 0 | + | 34 35 2 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 2 0 0 0 0 | + | 36 37 2 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 9 32 1 0 0 0 0 | + | 9 32 1 0 0 0 0 |
| − | 36 38 1 0 0 0 0 | + | 36 38 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 34 39 1 0 0 0 0 | + | 34 39 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 35 41 1 0 0 0 0 | + | 35 41 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 39 40 | + | M SAL 1 2 39 40 |
| − | M SBL 1 1 44 | + | M SBL 1 1 44 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 44 -1.1122 0.6422 | + | M SBV 1 44 -1.1122 0.6422 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 41 42 | + | M SAL 2 2 41 42 |
| − | M SBL 2 1 46 | + | M SBL 2 1 46 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SBV 2 46 -0.7525 -0.4345 | + | M SBV 2 46 -0.7525 -0.4345 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL3FAKCS0001 | + | ID FL3FAKCS0001 |
| − | FORMULA C27H30O15 | + | FORMULA C27H30O15 |
| − | EXACTMASS 594.15847029 | + | EXACTMASS 594.15847029 |
| − | AVERAGEMASS 594.5181 | + | AVERAGEMASS 594.5181 |
| − | SMILES C(C5O)(OCC(C5O)O)c(c(O)1)c(O)c(C(=O)3)c(OC(c(c4)cc(c(c(O)4)OC)OC)=C3)c(C(O2)C(O)C(O)C(O)C2)1 | + | SMILES C(C5O)(OCC(C5O)O)c(c(O)1)c(O)c(C(=O)3)c(OC(c(c4)cc(c(c(O)4)OC)OC)=C3)c(C(O2)C(O)C(O)C(O)C2)1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
42 46 0 0 0 0 0 0 0 0999 V2000
-2.3637 -0.6728 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3637 -1.4977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6493 -1.9102 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9348 -1.4977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9348 -0.6728 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6493 -0.2602 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2203 -1.9102 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4941 -1.4977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4941 -0.6728 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2203 -0.2602 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2203 -2.5535 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0779 -0.2604 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4844 2.5925 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2621 2.0033 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9321 1.1549 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9234 0.3361 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3282 0.9312 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7201 1.6734 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7544 2.5775 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3055 2.7348 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3551 0.6674 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6493 -2.7348 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.8308 -1.5266 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3541 -2.1559 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6678 -1.8889 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0053 -1.8817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4867 -1.4004 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0872 -1.7173 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.8317 -0.8394 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.0973 -2.1920 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0668 -2.3443 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3672 -0.1279 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1201 -0.5626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8729 -0.1279 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8729 0.7414 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1201 1.1761 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3672 0.7414 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1201 2.0450 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.9851 -0.7701 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.0973 -1.4124 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6254 1.1759 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.5430 0.6461 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
1 12 1 0 0 0 0
13 14 1 1 0 0 0
14 15 1 1 0 0 0
16 15 1 1 0 0 0
16 17 1 0 0 0 0
17 18 1 0 0 0 0
18 13 1 0 0 0 0
13 19 1 0 0 0 0
14 20 1 0 0 0 0
15 21 1 0 0 0 0
6 16 1 0 0 0 0
3 22 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 28 1 0 0 0 0
28 23 1 0 0 0 0
23 29 1 0 0 0 0
24 30 1 0 0 0 0
25 31 1 0 0 0 0
26 2 1 0 0 0 0
32 33 2 0 0 0 0
33 34 1 0 0 0 0
34 35 2 0 0 0 0
35 36 1 0 0 0 0
36 37 2 0 0 0 0
37 32 1 0 0 0 0
9 32 1 0 0 0 0
36 38 1 0 0 0 0
39 40 1 0 0 0 0
34 39 1 0 0 0 0
41 42 1 0 0 0 0
35 41 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 39 40
M SBL 1 1 44
M SMT 1 OCH3
M SBV 1 44 -1.1122 0.6422
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 41 42
M SBL 2 1 46
M SMT 2 OCH3
M SBV 2 46 -0.7525 -0.4345
S SKP 5
ID FL3FAKCS0001
FORMULA C27H30O15
EXACTMASS 594.15847029
AVERAGEMASS 594.5181
SMILES C(C5O)(OCC(C5O)O)c(c(O)1)c(O)c(C(=O)3)c(OC(c(c4)cc(c(c(O)4)OC)OC)=C3)c(C(O2)C(O)C(O)C(O)C2)1
M END