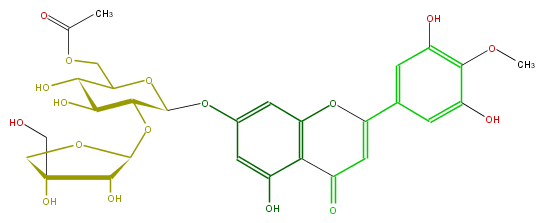
Mol:FL3FAJGS0001
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 46 50 0 0 0 0 0 0 0 0999 V2000 | + | 46 50 0 0 0 0 0 0 0 0999 V2000 |
| − | 2.8429 0.9987 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8429 0.9987 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8429 0.1737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8429 0.1737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5574 -0.2389 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5574 -0.2389 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2719 0.1737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2719 0.1737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2719 0.9987 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2719 0.9987 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5574 1.4113 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5574 1.4113 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1283 -0.2389 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1283 -0.2389 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4139 0.1737 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4139 0.1737 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6993 -0.2389 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6993 -0.2389 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6993 -1.0639 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6993 -1.0639 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4139 -1.4765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4139 -1.4765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1283 -1.0639 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1283 -1.0639 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0152 0.1737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0152 0.1737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7297 -0.2389 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7297 -0.2389 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7297 -1.0639 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7297 -1.0639 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0152 -1.4765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0152 -1.4765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4139 -2.1947 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4139 -2.1947 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4442 0.1737 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4442 0.1737 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0152 -2.1624 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0152 -2.1624 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9462 1.4068 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9462 1.4068 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5574 2.0660 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5574 2.0660 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8850 -0.1803 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8850 -0.1803 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.5711 1.0460 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.5711 1.0460 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2858 0.7220 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2858 0.7220 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8733 0.0074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8733 0.0074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0752 0.2165 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0752 0.2165 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2819 -0.0103 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2819 -0.0103 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6944 0.7044 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6944 0.7044 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4926 0.4954 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4926 0.4954 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7261 -0.3881 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7261 -0.3881 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5954 0.2009 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5954 0.2009 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9841 0.5349 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9841 0.5349 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8400 1.0562 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8400 1.0562 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4794 1.1387 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4794 1.1387 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0442 2.1454 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0442 2.1454 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4794 1.8193 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4794 1.8193 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8292 2.1947 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8292 2.1947 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9898 -1.4940 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9898 -1.4940 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9898 -1.9982 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9898 -1.9982 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5497 -1.9350 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5497 -1.9350 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9701 -0.5952 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9701 -0.5952 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1024 -1.0561 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1024 -1.0561 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.4340 -1.0561 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.4340 -1.0561 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5497 -1.4940 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5497 -1.4940 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2674 -0.7415 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2674 -0.7415 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.5711 -0.2482 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.5711 -0.2482 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 2 7 1 0 0 0 0 | + | 2 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 11 1 0 0 0 0 | + | 10 11 1 0 0 0 0 |
| − | 11 12 1 0 0 0 0 | + | 11 12 1 0 0 0 0 |
| − | 12 7 2 0 0 0 0 | + | 12 7 2 0 0 0 0 |
| − | 9 13 1 0 0 0 0 | + | 9 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 10 1 0 0 0 0 | + | 16 10 1 0 0 0 0 |
| − | 14 18 1 0 0 0 0 | + | 14 18 1 0 0 0 0 |
| − | 11 17 2 0 0 0 0 | + | 11 17 2 0 0 0 0 |
| − | 16 19 1 0 0 0 0 | + | 16 19 1 0 0 0 0 |
| − | 5 20 1 0 0 0 0 | + | 5 20 1 0 0 0 0 |
| − | 6 21 1 0 0 0 0 | + | 6 21 1 0 0 0 0 |
| − | 4 22 1 0 0 0 0 | + | 4 22 1 0 0 0 0 |
| − | 20 23 1 0 0 0 0 | + | 20 23 1 0 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 25 26 1 1 0 0 0 | + | 25 26 1 1 0 0 0 |
| − | 27 26 1 1 0 0 0 | + | 27 26 1 1 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 29 24 1 0 0 0 0 | + | 29 24 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 25 31 1 0 0 0 0 | + | 25 31 1 0 0 0 0 |
| − | 24 32 1 0 0 0 0 | + | 24 32 1 0 0 0 0 |
| − | 29 33 1 0 0 0 0 | + | 29 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 36 1 0 0 0 0 | + | 34 36 1 0 0 0 0 |
| − | 35 36 2 0 0 0 0 | + | 35 36 2 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 27 18 1 0 0 0 0 | + | 27 18 1 0 0 0 0 |
| − | 44 38 1 1 0 0 0 | + | 44 38 1 1 0 0 0 |
| − | 43 38 1 1 0 0 0 | + | 43 38 1 1 0 0 0 |
| − | 42 44 1 1 0 0 0 | + | 42 44 1 1 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 44 40 1 0 0 0 0 | + | 44 40 1 0 0 0 0 |
| − | 45 42 1 0 0 0 0 | + | 45 42 1 0 0 0 0 |
| − | 41 46 1 0 0 0 0 | + | 41 46 1 0 0 0 0 |
| − | 43 45 1 0 0 0 0 | + | 43 45 1 0 0 0 0 |
| − | 38 41 1 0 0 0 0 | + | 38 41 1 0 0 0 0 |
| − | 42 30 1 0 0 0 0 | + | 42 30 1 0 0 0 0 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL3FAJGS0001 | + | ID FL3FAJGS0001 |
| − | FORMULA C29H32O17 | + | FORMULA C29H32O17 |
| − | EXACTMASS 652.163949598 | + | EXACTMASS 652.163949598 |
| − | AVERAGEMASS 652.55418 | + | AVERAGEMASS 652.55418 |
| − | SMILES Oc(c1)c(OC)c(O)cc1C(=C5)Oc(c2)c(C(=O)5)c(O)cc2OC(C(OC(C4O)OCC4(CO)O)3)OC(C(C3O)O)COC(C)=O | + | SMILES Oc(c1)c(OC)c(O)cc1C(=C5)Oc(c2)c(C(=O)5)c(O)cc2OC(C(OC(C4O)OCC4(CO)O)3)OC(C(C3O)O)COC(C)=O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
46 50 0 0 0 0 0 0 0 0999 V2000
2.8429 0.9987 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8429 0.1737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5574 -0.2389 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2719 0.1737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2719 0.9987 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5574 1.4113 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1283 -0.2389 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4139 0.1737 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6993 -0.2389 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6993 -1.0639 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4139 -1.4765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1283 -1.0639 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0152 0.1737 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7297 -0.2389 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7297 -1.0639 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0152 -1.4765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4139 -2.1947 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4442 0.1737 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0152 -2.1624 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.9462 1.4068 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5574 2.0660 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.8850 -0.1803 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.5711 1.0460 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2858 0.7220 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8733 0.0074 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0752 0.2165 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2819 -0.0103 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6944 0.7044 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4926 0.4954 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7261 -0.3881 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.5954 0.2009 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.9841 0.5349 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.8400 1.0562 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4794 1.1387 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.0442 2.1454 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.4794 1.8193 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8292 2.1947 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.9898 -1.4940 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.9898 -1.9982 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5497 -1.9350 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.9701 -0.5952 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1024 -1.0561 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.4340 -1.0561 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5497 -1.4940 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2674 -0.7415 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.5711 -0.2482 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
2 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 11 1 0 0 0 0
11 12 1 0 0 0 0
12 7 2 0 0 0 0
9 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 10 1 0 0 0 0
14 18 1 0 0 0 0
11 17 2 0 0 0 0
16 19 1 0 0 0 0
5 20 1 0 0 0 0
6 21 1 0 0 0 0
4 22 1 0 0 0 0
20 23 1 0 0 0 0
24 25 1 1 0 0 0
25 26 1 1 0 0 0
27 26 1 1 0 0 0
27 28 1 0 0 0 0
28 29 1 0 0 0 0
29 24 1 0 0 0 0
26 30 1 0 0 0 0
25 31 1 0 0 0 0
24 32 1 0 0 0 0
29 33 1 0 0 0 0
33 34 1 0 0 0 0
34 36 1 0 0 0 0
35 36 2 0 0 0 0
36 37 1 0 0 0 0
27 18 1 0 0 0 0
44 38 1 1 0 0 0
43 38 1 1 0 0 0
42 44 1 1 0 0 0
38 39 1 0 0 0 0
44 40 1 0 0 0 0
45 42 1 0 0 0 0
41 46 1 0 0 0 0
43 45 1 0 0 0 0
38 41 1 0 0 0 0
42 30 1 0 0 0 0
S SKP 5
ID FL3FAJGS0001
FORMULA C29H32O17
EXACTMASS 652.163949598
AVERAGEMASS 652.55418
SMILES Oc(c1)c(OC)c(O)cc1C(=C5)Oc(c2)c(C(=O)5)c(O)cc2OC(C(OC(C4O)OCC4(CO)O)3)OC(C(C3O)O)COC(C)=O
M END