Mol:FL3FAGGS0004
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 46 50 0 0 0 0 0 0 0 0999 V2000 | + | 46 50 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.4419 -1.0622 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4419 -1.0622 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4419 -1.9315 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4419 -1.9315 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7444 -2.3343 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7444 -2.3343 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0467 -1.9315 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0467 -1.9315 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0467 -1.1259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0467 -1.1259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7444 -0.7231 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7444 -0.7231 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3490 -2.3343 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3490 -2.3343 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6514 -1.9315 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6514 -1.9315 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6514 -1.1259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6514 -1.1259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3490 -0.7231 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3490 -0.7231 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3490 -3.0442 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3490 -3.0442 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0459 -0.7233 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0459 -0.7233 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7571 -1.1338 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7571 -1.1338 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4681 -0.7233 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4681 -0.7233 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4681 0.0977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4681 0.0977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7571 0.5083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7571 0.5083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0459 0.0977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0459 0.0977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7430 -3.0157 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7430 -3.0157 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1548 -0.6744 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1548 -0.6744 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7571 1.2708 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7571 1.2708 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1789 0.5081 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1789 0.5081 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1789 -1.1338 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1789 -1.1338 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2326 -0.4001 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2326 -0.4001 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7531 -1.2308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7531 -1.2308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6753 -0.9673 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6753 -0.9673 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6033 -1.2308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6033 -1.2308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0832 -0.4001 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0832 -0.4001 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1606 -0.6636 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1606 -0.6636 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5594 -0.5461 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5594 -0.5461 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7275 -0.0071 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7275 -0.0071 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.8591 -0.5535 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.8591 -0.5535 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6755 0.5928 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6755 0.5928 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8982 0.1440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8982 0.1440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1448 1.0071 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1448 1.0071 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8982 1.8753 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8982 1.8753 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6755 2.3242 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6755 2.3242 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4289 1.4610 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4289 1.4610 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3020 2.9630 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3020 2.9630 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8542 2.4738 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8542 2.4738 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5577 0.6681 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5577 0.6681 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0486 2.0234 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0486 2.0234 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0486 3.0442 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0486 3.0442 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.9327 1.5130 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.9327 1.5130 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.4223 -1.0259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.4223 -1.0259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.9327 -0.1419 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.9327 -0.1419 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.9327 -1.9100 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.9327 -1.9100 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 16 20 1 0 0 0 0 | + | 16 20 1 0 0 0 0 |
| − | 15 21 1 0 0 0 0 | + | 15 21 1 0 0 0 0 |
| − | 14 22 1 0 0 0 0 | + | 14 22 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 25 26 1 1 0 0 0 | + | 25 26 1 1 0 0 0 |
| − | 27 26 1 1 0 0 0 | + | 27 26 1 1 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 23 1 0 0 0 0 | + | 28 23 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 28 30 1 0 0 0 0 | + | 28 30 1 0 0 0 0 |
| − | 27 31 1 0 0 0 0 | + | 27 31 1 0 0 0 0 |
| − | 24 22 1 0 0 0 0 | + | 24 22 1 0 0 0 0 |
| − | 33 32 1 1 0 0 0 | + | 33 32 1 1 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 1 1 0 0 0 | + | 36 37 1 1 0 0 0 |
| − | 37 32 1 1 0 0 0 | + | 37 32 1 1 0 0 0 |
| − | 36 38 1 0 0 0 0 | + | 36 38 1 0 0 0 0 |
| − | 35 39 1 0 0 0 0 | + | 35 39 1 0 0 0 0 |
| − | 34 40 1 0 0 0 0 | + | 34 40 1 0 0 0 0 |
| − | 33 19 1 0 0 0 0 | + | 33 19 1 0 0 0 0 |
| − | 41 42 2 0 0 0 0 | + | 41 42 2 0 0 0 0 |
| − | 41 43 1 0 0 0 0 | + | 41 43 1 0 0 0 0 |
| − | 37 41 1 0 0 0 0 | + | 37 41 1 0 0 0 0 |
| − | 44 45 2 0 0 0 0 | + | 44 45 2 0 0 0 0 |
| − | 44 46 1 0 0 0 0 | + | 44 46 1 0 0 0 0 |
| − | 26 44 1 0 0 0 0 | + | 26 44 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 3 41 42 43 | + | M SAL 1 3 41 42 43 |
| − | M SBL 1 1 47 | + | M SBL 1 1 47 |
| − | M SMT 1 ^COOH | + | M SMT 1 ^COOH |
| − | M SBV 1 47 0.6197 -0.5624 | + | M SBV 1 47 0.6197 -0.5624 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 3 44 45 46 | + | M SAL 2 3 44 45 46 |
| − | M SBL 2 1 50 | + | M SBL 2 1 50 |
| − | M SMT 2 COOH | + | M SMT 2 COOH |
| − | M SBV 2 50 -0.8190 -0.2049 | + | M SBV 2 50 -0.8190 -0.2049 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL3FAGGS0004 | + | ID FL3FAGGS0004 |
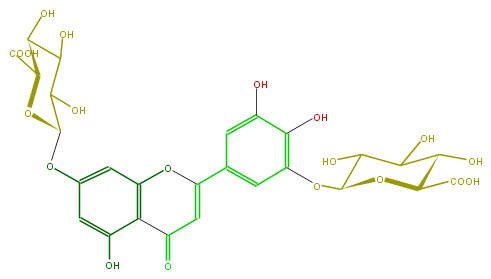
| − | FORMULA C27H26O19 | + | FORMULA C27H26O19 |
| − | EXACTMASS 654.1068286499999 | + | EXACTMASS 654.1068286499999 |
| − | AVERAGEMASS 654.4839400000001 | + | AVERAGEMASS 654.4839400000001 |
| − | SMILES c(c2)(C(=C3)Oc(c4)c(c(cc4OC(O5)C(C(C(O)C5C(O)=O)O)O)O)C3=O)cc(c(c2O)O)OC(C(O)1)OC(C(O)=O)C(O)C(O)1 | + | SMILES c(c2)(C(=C3)Oc(c4)c(c(cc4OC(O5)C(C(C(O)C5C(O)=O)O)O)O)C3=O)cc(c(c2O)O)OC(C(O)1)OC(C(O)=O)C(O)C(O)1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
46 50 0 0 0 0 0 0 0 0999 V2000
-3.4419 -1.0622 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4419 -1.9315 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7444 -2.3343 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0467 -1.9315 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0467 -1.1259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7444 -0.7231 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3490 -2.3343 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6514 -1.9315 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6514 -1.1259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3490 -0.7231 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3490 -3.0442 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0459 -0.7233 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7571 -1.1338 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4681 -0.7233 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4681 0.0977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7571 0.5083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0459 0.0977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7430 -3.0157 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1548 -0.6744 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7571 1.2708 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1789 0.5081 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1789 -1.1338 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2326 -0.4001 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7531 -1.2308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6753 -0.9673 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.6033 -1.2308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.0832 -0.4001 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1606 -0.6636 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5594 -0.5461 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.7275 -0.0071 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.8591 -0.5535 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.6755 0.5928 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.8982 0.1440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1448 1.0071 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8982 1.8753 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.6755 2.3242 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4289 1.4610 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3020 2.9630 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.8542 2.4738 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5577 0.6681 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.0486 2.0234 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.0486 3.0442 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.9327 1.5130 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.4223 -1.0259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.9327 -0.1419 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.9327 -1.9100 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
1 19 1 0 0 0 0
16 20 1 0 0 0 0
15 21 1 0 0 0 0
14 22 1 0 0 0 0
23 24 1 0 0 0 0
24 25 1 1 0 0 0
25 26 1 1 0 0 0
27 26 1 1 0 0 0
27 28 1 0 0 0 0
28 23 1 0 0 0 0
23 29 1 0 0 0 0
28 30 1 0 0 0 0
27 31 1 0 0 0 0
24 22 1 0 0 0 0
33 32 1 1 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 37 1 1 0 0 0
37 32 1 1 0 0 0
36 38 1 0 0 0 0
35 39 1 0 0 0 0
34 40 1 0 0 0 0
33 19 1 0 0 0 0
41 42 2 0 0 0 0
41 43 1 0 0 0 0
37 41 1 0 0 0 0
44 45 2 0 0 0 0
44 46 1 0 0 0 0
26 44 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 3 41 42 43
M SBL 1 1 47
M SMT 1 ^COOH
M SBV 1 47 0.6197 -0.5624
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 3 44 45 46
M SBL 2 1 50
M SMT 2 COOH
M SBV 2 50 -0.8190 -0.2049
S SKP 5
ID FL3FAGGS0004
FORMULA C27H26O19
EXACTMASS 654.1068286499999
AVERAGEMASS 654.4839400000001
SMILES c(c2)(C(=C3)Oc(c4)c(c(cc4OC(O5)C(C(C(O)C5C(O)=O)O)O)O)C3=O)cc(c(c2O)O)OC(C(O)1)OC(C(O)=O)C(O)C(O)1
M END