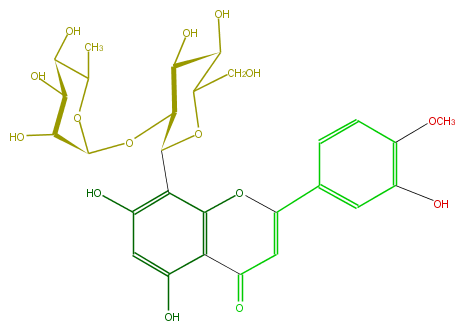
Mol:FL3FAECS0004
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 43 47 0 0 0 0 0 0 0 0999 V2000 | + | 43 47 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.3308 -0.9677 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3308 -0.9677 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3308 -1.7927 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3308 -1.7927 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6164 -2.2052 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6164 -2.2052 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9020 -1.7927 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9020 -1.7927 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9020 -0.9677 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9020 -0.9677 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6164 -0.5554 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6164 -0.5554 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1876 -2.2052 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1876 -2.2052 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5269 -1.7927 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5269 -1.7927 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5269 -0.9677 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5269 -0.9677 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1876 -0.5554 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1876 -0.5554 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1876 -2.8484 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1876 -2.8484 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0450 -0.5555 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0450 -0.5555 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5903 2.2473 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5903 2.2473 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5227 1.9596 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5227 1.9596 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5028 1.0494 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5028 1.0494 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7745 0.2772 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7745 0.2772 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0118 0.6326 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0118 0.6326 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1261 1.4642 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1261 1.4642 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6217 3.0298 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6217 3.0298 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2464 2.6482 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2464 2.6482 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3933 0.4579 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3933 0.4579 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6164 -3.0298 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6164 -3.0298 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4000 -0.4229 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4000 -0.4229 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1528 -0.8577 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1528 -0.8577 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9057 -0.4229 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9057 -0.4229 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9057 0.4464 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9057 0.4464 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1528 0.8809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1528 0.8809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4000 0.4464 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4000 0.4464 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7728 -0.7607 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7728 -0.7607 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9041 0.5994 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9041 0.5994 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2296 0.2099 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2296 0.2099 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4436 0.9588 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4436 0.9588 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2296 1.7122 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2296 1.7122 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9041 2.1017 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9041 2.1017 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6902 1.3527 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6902 1.3527 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5962 2.6957 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5962 2.6957 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2283 2.3618 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2283 2.3618 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2955 1.7821 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2955 1.7821 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5757 0.5691 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5757 0.5691 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3560 1.8481 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3560 1.8481 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3250 1.0365 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3250 1.0365 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6581 0.8807 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6581 0.8807 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5757 0.3509 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5757 0.3509 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 1 12 1 0 0 0 0 | + | 1 12 1 0 0 0 0 |
| − | 13 14 1 1 0 0 0 | + | 13 14 1 1 0 0 0 |
| − | 14 15 1 1 0 0 0 | + | 14 15 1 1 0 0 0 |
| − | 16 15 1 1 0 0 0 | + | 16 15 1 1 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 18 1 0 0 0 0 | + | 17 18 1 0 0 0 0 |
| − | 18 13 1 0 0 0 0 | + | 18 13 1 0 0 0 0 |
| − | 13 19 1 0 0 0 0 | + | 13 19 1 0 0 0 0 |
| − | 14 20 1 0 0 0 0 | + | 14 20 1 0 0 0 0 |
| − | 15 21 1 0 0 0 0 | + | 15 21 1 0 0 0 0 |
| − | 6 16 1 0 0 0 0 | + | 6 16 1 0 0 0 0 |
| − | 3 22 1 0 0 0 0 | + | 3 22 1 0 0 0 0 |
| − | 23 24 2 0 0 0 0 | + | 23 24 2 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 2 0 0 0 0 | + | 25 26 2 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 2 0 0 0 0 | + | 27 28 2 0 0 0 0 |
| − | 28 23 1 0 0 0 0 | + | 28 23 1 0 0 0 0 |
| − | 9 23 1 0 0 0 0 | + | 9 23 1 0 0 0 0 |
| − | 25 29 1 0 0 0 0 | + | 25 29 1 0 0 0 0 |
| − | 31 30 1 1 0 0 0 | + | 31 30 1 1 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 1 0 0 0 | + | 34 35 1 1 0 0 0 |
| − | 35 30 1 1 0 0 0 | + | 35 30 1 1 0 0 0 |
| − | 34 36 1 0 0 0 0 | + | 34 36 1 0 0 0 0 |
| − | 33 37 1 0 0 0 0 | + | 33 37 1 0 0 0 0 |
| − | 35 38 1 0 0 0 0 | + | 35 38 1 0 0 0 0 |
| − | 30 39 1 0 0 0 0 | + | 30 39 1 0 0 0 0 |
| − | 31 21 1 0 0 0 0 | + | 31 21 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 18 40 1 0 0 0 0 | + | 18 40 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 26 42 1 0 0 0 0 | + | 26 42 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 40 41 | + | M SAL 1 2 40 41 |
| − | M SBL 1 1 45 | + | M SBL 1 1 45 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 45 -0.7701 -0.3840 | + | M SBV 1 45 -0.7701 -0.3840 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 42 43 | + | M SAL 2 2 42 43 |
| − | M SBL 2 1 47 | + | M SBL 2 1 47 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SBV 2 47 -0.7524 -0.4343 | + | M SBV 2 47 -0.7524 -0.4343 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL3FAECS0004 | + | ID FL3FAECS0004 |
| − | FORMULA C28H32O15 | + | FORMULA C28H32O15 |
| − | EXACTMASS 608.174120354 | + | EXACTMASS 608.174120354 |
| − | AVERAGEMASS 608.54468 | + | AVERAGEMASS 608.54468 |
| − | SMILES OC(C4O)C(OC(O5)C(O)C(O)C(C(C)5)O)C(OC(CO)4)c(c12)c(cc(O)c1C(=O)C=C(c(c3)ccc(OC)c3O)O2)O | + | SMILES OC(C4O)C(OC(O5)C(O)C(O)C(C(C)5)O)C(OC(CO)4)c(c12)c(cc(O)c1C(=O)C=C(c(c3)ccc(OC)c3O)O2)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
43 47 0 0 0 0 0 0 0 0999 V2000
-2.3308 -0.9677 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3308 -1.7927 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6164 -2.2052 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9020 -1.7927 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9020 -0.9677 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6164 -0.5554 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1876 -2.2052 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5269 -1.7927 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5269 -0.9677 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1876 -0.5554 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1876 -2.8484 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0450 -0.5555 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5903 2.2473 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5227 1.9596 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5028 1.0494 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7745 0.2772 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0118 0.6326 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.1261 1.4642 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6217 3.0298 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2464 2.6482 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3933 0.4579 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6164 -3.0298 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4000 -0.4229 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1528 -0.8577 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9057 -0.4229 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9057 0.4464 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1528 0.8809 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4000 0.4464 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7728 -0.7607 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9041 0.5994 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2296 0.2099 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4436 0.9588 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2296 1.7122 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9041 2.1017 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6902 1.3527 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5962 2.6957 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2283 2.3618 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2955 1.7821 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.5757 0.5691 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.3560 1.8481 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3250 1.0365 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6581 0.8807 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.5757 0.3509 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
1 12 1 0 0 0 0
13 14 1 1 0 0 0
14 15 1 1 0 0 0
16 15 1 1 0 0 0
16 17 1 0 0 0 0
17 18 1 0 0 0 0
18 13 1 0 0 0 0
13 19 1 0 0 0 0
14 20 1 0 0 0 0
15 21 1 0 0 0 0
6 16 1 0 0 0 0
3 22 1 0 0 0 0
23 24 2 0 0 0 0
24 25 1 0 0 0 0
25 26 2 0 0 0 0
26 27 1 0 0 0 0
27 28 2 0 0 0 0
28 23 1 0 0 0 0
9 23 1 0 0 0 0
25 29 1 0 0 0 0
31 30 1 1 0 0 0
31 32 1 0 0 0 0
32 33 1 0 0 0 0
33 34 1 0 0 0 0
34 35 1 1 0 0 0
35 30 1 1 0 0 0
34 36 1 0 0 0 0
33 37 1 0 0 0 0
35 38 1 0 0 0 0
30 39 1 0 0 0 0
31 21 1 0 0 0 0
40 41 1 0 0 0 0
18 40 1 0 0 0 0
42 43 1 0 0 0 0
26 42 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 40 41
M SBL 1 1 45
M SMT 1 CH2OH
M SBV 1 45 -0.7701 -0.3840
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 42 43
M SBL 2 1 47
M SMT 2 OCH3
M SBV 2 47 -0.7524 -0.4343
S SKP 5
ID FL3FAECS0004
FORMULA C28H32O15
EXACTMASS 608.174120354
AVERAGEMASS 608.54468
SMILES OC(C4O)C(OC(O5)C(O)C(O)C(C(C)5)O)C(OC(CO)4)c(c12)c(cc(O)c1C(=O)C=C(c(c3)ccc(OC)c3O)O2)O
M END