Mol:FL3FADGS0020
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 44 48 0 0 0 0 0 0 0 0999 V2000 | + | 44 48 0 0 0 0 0 0 0 0999 V2000 |
| − | 0.5664 1.5048 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5664 1.5048 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5664 0.9840 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5664 0.9840 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0174 0.7236 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0174 0.7236 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4685 0.9840 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4685 0.9840 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4685 1.5048 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4685 1.5048 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0174 1.7652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0174 1.7652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9196 0.7236 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9196 0.7236 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3706 0.9840 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3706 0.9840 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3706 1.5048 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3706 1.5048 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9196 1.7652 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9196 1.7652 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0913 0.3557 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0913 0.3557 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8215 1.7652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8215 1.7652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2812 1.4997 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2812 1.4997 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7410 1.7652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7410 1.7652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7410 2.2960 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7410 2.2960 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2812 2.5614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2812 2.5614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8215 2.2960 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8215 2.2960 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7410 2.2960 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7410 2.2960 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1162 1.7647 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1162 1.7647 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5232 1.5768 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -2.5232 1.5768 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -2.0076 0.8962 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.0076 0.8962 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.2651 1.1850 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.2651 1.1850 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -0.5486 1.1927 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -0.5486 1.1927 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.0692 1.7134 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0692 1.7134 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7188 1.3706 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -1.7188 1.3706 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -3.2370 1.1647 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2370 1.1647 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8114 0.8571 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8114 0.8571 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8396 0.4708 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8396 0.4708 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1460 2.1105 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1460 2.1105 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0174 0.2038 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0174 0.2038 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5574 2.1105 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5574 2.1105 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3775 0.2910 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3775 0.2910 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8950 0.4296 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8950 0.4296 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3775 -0.4032 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3775 -0.4032 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8170 -0.6570 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8170 -0.6570 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8170 -1.3313 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8170 -1.3313 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3312 -1.6281 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3312 -1.6281 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3312 -2.2218 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3312 -2.2218 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8170 -2.5186 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8170 -2.5186 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3029 -2.2218 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3029 -2.2218 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3029 -1.6281 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3029 -1.6281 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8170 -3.1066 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8170 -3.1066 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5111 3.1066 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5111 3.1066 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8996 4.0280 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8996 4.0280 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 18 15 1 0 0 0 0 | + | 18 15 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 20 1 0 0 0 0 | + | 25 20 1 0 0 0 0 |
| − | 20 26 1 0 0 0 0 | + | 20 26 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 19 1 0 0 0 0 | + | 23 19 1 0 0 0 0 |
| − | 25 29 1 0 0 0 0 | + | 25 29 1 0 0 0 0 |
| − | 3 30 1 0 0 0 0 | + | 3 30 1 0 0 0 0 |
| − | 29 31 1 0 0 0 0 | + | 29 31 1 0 0 0 0 |
| − | 27 32 1 0 0 0 0 | + | 27 32 1 0 0 0 0 |
| − | 32 33 2 0 0 0 0 | + | 32 33 2 0 0 0 0 |
| − | 32 34 1 0 0 0 0 | + | 32 34 1 0 0 0 0 |
| − | 34 35 2 0 0 0 0 | + | 34 35 2 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 2 0 0 0 0 | + | 36 37 2 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 2 0 0 0 0 | + | 38 39 2 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 40 41 2 0 0 0 0 | + | 40 41 2 0 0 0 0 |
| − | 41 36 1 0 0 0 0 | + | 41 36 1 0 0 0 0 |
| − | 39 42 1 0 0 0 0 | + | 39 42 1 0 0 0 0 |
| − | 16 43 1 0 0 0 0 | + | 16 43 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 29 31 | + | M SAL 2 2 29 31 |
| − | M SBL 2 1 32 | + | M SBL 2 1 32 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SVB 2 32 -2.146 2.1105 | + | M SVB 2 32 -2.146 2.1105 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 43 44 | + | M SAL 1 2 43 44 |
| − | M SBL 1 1 47 | + | M SBL 1 1 47 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 47 3.5111 3.1066 | + | M SVB 1 47 3.5111 3.1066 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL3FADGS0020 | + | ID FL3FADGS0020 |
| − | KNApSAcK_ID C00004354 | + | KNApSAcK_ID C00004354 |
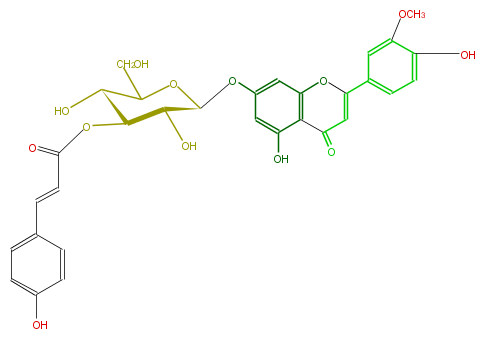
| − | NAME Luteolin 3'-methyl ether 7-(3''-E-p-coumarylglucoside) | + | NAME Luteolin 3'-methyl ether 7-(3''-E-p-coumarylglucoside) |
| − | CAS_RN 103450-98-2 | + | CAS_RN 103450-98-2 |
| − | FORMULA C31H28O13 | + | FORMULA C31H28O13 |
| − | EXACTMASS 608.152990982 | + | EXACTMASS 608.152990982 |
| − | AVERAGEMASS 608.5462200000001 | + | AVERAGEMASS 608.5462200000001 |
| − | SMILES C(CO)(O2)[C@@H]([C@@H]([C@@H]([C@@H]2Oc(c5)cc(O)c(c53)C(C=C(c(c4)cc(c(O)c4)OC)O3)=O)O)OC(=O)C=Cc(c1)ccc(O)c1)O | + | SMILES C(CO)(O2)[C@@H]([C@@H]([C@@H]([C@@H]2Oc(c5)cc(O)c(c53)C(C=C(c(c4)cc(c(O)c4)OC)O3)=O)O)OC(=O)C=Cc(c1)ccc(O)c1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
44 48 0 0 0 0 0 0 0 0999 V2000
0.5664 1.5048 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5664 0.9840 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0174 0.7236 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4685 0.9840 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4685 1.5048 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0174 1.7652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9196 0.7236 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3706 0.9840 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3706 1.5048 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9196 1.7652 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0913 0.3557 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8215 1.7652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2812 1.4997 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7410 1.7652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7410 2.2960 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2812 2.5614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8215 2.2960 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.7410 2.2960 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1162 1.7647 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5232 1.5768 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-2.0076 0.8962 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.2651 1.1850 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.5486 1.1927 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.0692 1.7134 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7188 1.3706 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-3.2370 1.1647 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8114 0.8571 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8396 0.4708 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1460 2.1105 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0174 0.2038 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5574 2.1105 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3775 0.2910 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8950 0.4296 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3775 -0.4032 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8170 -0.6570 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8170 -1.3313 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3312 -1.6281 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3312 -2.2218 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8170 -2.5186 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3029 -2.2218 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3029 -1.6281 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8170 -3.1066 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5111 3.1066 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.8996 4.0280 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
18 15 1 0 0 0 0
1 19 1 0 0 0 0
20 21 1 1 0 0 0
21 22 1 1 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 20 1 0 0 0 0
20 26 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
23 19 1 0 0 0 0
25 29 1 0 0 0 0
3 30 1 0 0 0 0
29 31 1 0 0 0 0
27 32 1 0 0 0 0
32 33 2 0 0 0 0
32 34 1 0 0 0 0
34 35 2 0 0 0 0
35 36 1 0 0 0 0
36 37 2 0 0 0 0
37 38 1 0 0 0 0
38 39 2 0 0 0 0
39 40 1 0 0 0 0
40 41 2 0 0 0 0
41 36 1 0 0 0 0
39 42 1 0 0 0 0
16 43 1 0 0 0 0
43 44 1 0 0 0 0
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 29 31
M SBL 2 1 32
M SMT 2 CH2OH
M SVB 2 32 -2.146 2.1105
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 43 44
M SBL 1 1 47
M SMT 1 OCH3
M SVB 1 47 3.5111 3.1066
S SKP 8
ID FL3FADGS0020
KNApSAcK_ID C00004354
NAME Luteolin 3'-methyl ether 7-(3''-E-p-coumarylglucoside)
CAS_RN 103450-98-2
FORMULA C31H28O13
EXACTMASS 608.152990982
AVERAGEMASS 608.5462200000001
SMILES C(CO)(O2)[C@@H]([C@@H]([C@@H]([C@@H]2Oc(c5)cc(O)c(c53)C(C=C(c(c4)cc(c(O)c4)OC)O3)=O)O)OC(=O)C=Cc(c1)ccc(O)c1)O
M END