Mol:FL3FACDS0010
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 53 58 0 0 0 0 0 0 0 0999 V2000 | + | 53 58 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.9219 -0.9253 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9219 -0.9253 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9219 -1.7503 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9219 -1.7503 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2075 -2.1629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2075 -2.1629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4930 -1.7503 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4930 -1.7503 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4930 -0.9253 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4930 -0.9253 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2075 -0.5129 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2075 -0.5129 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7785 -2.1629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7785 -2.1629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0641 -1.7503 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0641 -1.7503 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0641 -0.9253 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0641 -0.9253 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7785 -0.5129 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7785 -0.5129 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7785 -2.8060 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7785 -2.8060 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6361 -0.5130 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6361 -0.5130 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2075 -2.9875 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2075 -2.9875 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1910 -0.3806 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1910 -0.3806 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5619 -0.8152 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5619 -0.8152 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3148 -0.3806 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3148 -0.3806 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3148 0.4887 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3148 0.4887 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5619 0.9235 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5619 0.9235 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1910 0.4887 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1910 0.4887 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0672 0.9232 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0672 0.9232 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0672 -0.8150 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0672 -0.8150 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9042 2.2778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9042 2.2778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6820 1.6886 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6820 1.6886 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3521 0.8402 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3521 0.8402 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3432 0.0214 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3432 0.0214 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7482 0.6164 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7482 0.6164 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1399 1.3586 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1399 1.3586 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2771 2.9875 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2771 2.9875 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7047 2.5439 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7047 2.5439 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8368 0.2083 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8368 0.2083 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.2424 0.3176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.2424 0.3176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5679 -0.0720 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5679 -0.0720 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7819 0.6769 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7819 0.6769 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5679 1.4304 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5679 1.4304 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.2424 1.8199 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.2424 1.8199 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0285 1.0709 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0285 1.0709 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0788 2.3727 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0788 2.3727 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5679 1.9164 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5679 1.9164 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.6132 1.5003 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.6132 1.5003 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.7903 0.2872 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.7903 0.2872 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0458 1.5004 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0458 1.5004 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6595 0.8312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6595 0.8312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4023 1.0436 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4023 1.0436 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1498 0.8312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1498 0.8312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5362 1.5004 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5362 1.5004 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7933 1.2881 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7933 1.2881 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3915 1.3893 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3915 1.3893 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3274 1.8990 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3274 1.8990 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1364 1.3628 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1364 1.3628 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4914 2.0998 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4914 2.0998 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5737 1.5701 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5737 1.5701 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8726 0.9053 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8726 0.9053 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.7903 0.3756 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.7903 0.3756 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 1 12 1 0 0 0 0 | + | 1 12 1 0 0 0 0 |
| − | 3 13 1 0 0 0 0 | + | 3 13 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 18 1 0 0 0 0 | + | 17 18 1 0 0 0 0 |
| − | 18 19 2 0 0 0 0 | + | 18 19 2 0 0 0 0 |
| − | 19 14 1 0 0 0 0 | + | 19 14 1 0 0 0 0 |
| − | 9 14 1 0 0 0 0 | + | 9 14 1 0 0 0 0 |
| − | 17 20 1 0 0 0 0 | + | 17 20 1 0 0 0 0 |
| − | 16 21 1 0 0 0 0 | + | 16 21 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 6 25 1 0 0 0 0 | + | 6 25 1 0 0 0 0 |
| − | 32 31 1 1 0 0 0 | + | 32 31 1 1 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 1 0 0 0 | + | 35 36 1 1 0 0 0 |
| − | 36 31 1 1 0 0 0 | + | 36 31 1 1 0 0 0 |
| − | 35 37 1 0 0 0 0 | + | 35 37 1 0 0 0 0 |
| − | 34 38 1 0 0 0 0 | + | 34 38 1 0 0 0 0 |
| − | 36 39 1 0 0 0 0 | + | 36 39 1 0 0 0 0 |
| − | 31 40 1 0 0 0 0 | + | 31 40 1 0 0 0 0 |
| − | 32 30 1 0 0 0 0 | + | 32 30 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 42 43 1 1 0 0 0 | + | 42 43 1 1 0 0 0 |
| − | 43 44 1 1 0 0 0 | + | 43 44 1 1 0 0 0 |
| − | 45 44 1 1 0 0 0 | + | 45 44 1 1 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | 46 41 1 0 0 0 0 | + | 46 41 1 0 0 0 0 |
| − | 41 47 1 0 0 0 0 | + | 41 47 1 0 0 0 0 |
| − | 46 48 1 0 0 0 0 | + | 46 48 1 0 0 0 0 |
| − | 45 49 1 0 0 0 0 | + | 45 49 1 0 0 0 0 |
| − | 42 20 1 0 0 0 0 | + | 42 20 1 0 0 0 0 |
| − | 50 51 1 0 0 0 0 | + | 50 51 1 0 0 0 0 |
| − | 27 50 1 0 0 0 0 | + | 27 50 1 0 0 0 0 |
| − | 52 53 1 0 0 0 0 | + | 52 53 1 0 0 0 0 |
| − | 44 52 1 0 0 0 0 | + | 44 52 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 50 51 | + | M SAL 1 2 50 51 |
| − | M SBL 1 1 56 | + | M SBL 1 1 56 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 56 -0.6485 -0.7412 | + | M SBV 1 56 -0.6485 -0.7412 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 52 53 | + | M SAL 2 2 52 53 |
| − | M SBL 2 1 58 | + | M SBL 2 1 58 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SBV 2 58 -0.7229 -0.0742 | + | M SBV 2 58 -0.7229 -0.0742 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL3FACDS0010 | + | ID FL3FACDS0010 |
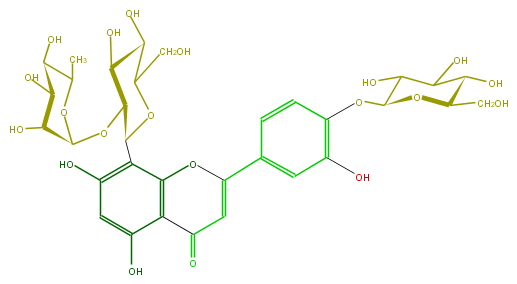
| − | FORMULA C33H40O20 | + | FORMULA C33H40O20 |
| − | EXACTMASS 756.21129372 | + | EXACTMASS 756.21129372 |
| − | AVERAGEMASS 756.6587 | + | AVERAGEMASS 756.6587 |
| − | SMILES C(=C(c(c5)cc(c(OC(O6)C(O)C(C(C6CO)O)O)c5)O)4)C(c(c1O4)c(O)cc(c(C(C2OC(C3O)OC(C(C(O)3)O)C)OC(CO)C(C2O)O)1)O)=O | + | SMILES C(=C(c(c5)cc(c(OC(O6)C(O)C(C(C6CO)O)O)c5)O)4)C(c(c1O4)c(O)cc(c(C(C2OC(C3O)OC(C(C(O)3)O)C)OC(CO)C(C2O)O)1)O)=O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
53 58 0 0 0 0 0 0 0 0999 V2000
-3.9219 -0.9253 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9219 -1.7503 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2075 -2.1629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4930 -1.7503 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4930 -0.9253 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2075 -0.5129 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7785 -2.1629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0641 -1.7503 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0641 -0.9253 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7785 -0.5129 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7785 -2.8060 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.6361 -0.5130 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2075 -2.9875 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1910 -0.3806 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5619 -0.8152 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3148 -0.3806 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3148 0.4887 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5619 0.9235 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1910 0.4887 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0672 0.9232 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0672 -0.8150 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9042 2.2778 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6820 1.6886 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3521 0.8402 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3432 0.0214 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7482 0.6164 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1399 1.3586 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2771 2.9875 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7047 2.5439 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.8368 0.2083 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.2424 0.3176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.5679 -0.0720 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.7819 0.6769 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.5679 1.4304 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.2424 1.8199 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.0285 1.0709 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.0788 2.3727 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.5679 1.9164 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.6132 1.5003 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.7903 0.2872 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0458 1.5004 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6595 0.8312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4023 1.0436 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1498 0.8312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5362 1.5004 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7933 1.2881 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3915 1.3893 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.3274 1.8990 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.1364 1.3628 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4914 2.0998 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5737 1.5701 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.8726 0.9053 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.7903 0.3756 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
1 12 1 0 0 0 0
3 13 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 18 1 0 0 0 0
18 19 2 0 0 0 0
19 14 1 0 0 0 0
9 14 1 0 0 0 0
17 20 1 0 0 0 0
16 21 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
24 30 1 0 0 0 0
6 25 1 0 0 0 0
32 31 1 1 0 0 0
32 33 1 0 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 36 1 1 0 0 0
36 31 1 1 0 0 0
35 37 1 0 0 0 0
34 38 1 0 0 0 0
36 39 1 0 0 0 0
31 40 1 0 0 0 0
32 30 1 0 0 0 0
41 42 1 0 0 0 0
42 43 1 1 0 0 0
43 44 1 1 0 0 0
45 44 1 1 0 0 0
45 46 1 0 0 0 0
46 41 1 0 0 0 0
41 47 1 0 0 0 0
46 48 1 0 0 0 0
45 49 1 0 0 0 0
42 20 1 0 0 0 0
50 51 1 0 0 0 0
27 50 1 0 0 0 0
52 53 1 0 0 0 0
44 52 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 50 51
M SBL 1 1 56
M SMT 1 CH2OH
M SBV 1 56 -0.6485 -0.7412
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 52 53
M SBL 2 1 58
M SMT 2 CH2OH
M SBV 2 58 -0.7229 -0.0742
S SKP 5
ID FL3FACDS0010
FORMULA C33H40O20
EXACTMASS 756.21129372
AVERAGEMASS 756.6587
SMILES C(=C(c(c5)cc(c(OC(O6)C(O)C(C(C6CO)O)O)c5)O)4)C(c(c1O4)c(O)cc(c(C(C2OC(C3O)OC(C(C(O)3)O)C)OC(CO)C(C2O)O)1)O)=O
M END