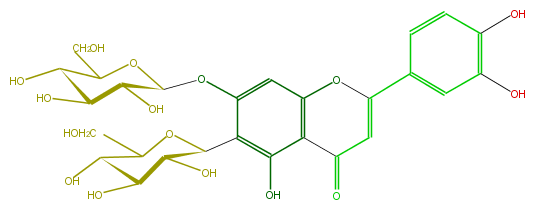
Mol:FL3FACDS0006
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 43 47 0 0 0 0 0 0 0 0999 V2000 | + | 43 47 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.5959 0.1120 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5959 0.1120 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5959 -0.7063 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5959 -0.7063 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1127 -1.1153 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1127 -1.1153 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8213 -0.7063 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8213 -0.7063 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8213 0.1120 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8213 0.1120 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1127 0.5212 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1127 0.5212 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5299 -1.1153 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5299 -1.1153 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2385 -0.7063 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2385 -0.7063 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2385 0.1120 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2385 0.1120 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5299 0.5212 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5299 0.5212 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5299 -1.9456 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5299 -1.9456 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1127 -1.9333 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1127 -1.9333 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1045 0.6524 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1045 0.6524 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8512 0.2212 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8512 0.2212 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5979 0.6524 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5979 0.6524 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5979 1.5146 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5979 1.5146 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8512 1.9456 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8512 1.9456 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1045 1.5146 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1045 1.5146 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.3442 1.9454 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.3442 1.9454 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3494 0.5425 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3494 0.5425 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5134 -1.1523 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5134 -1.1523 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6885 -1.6928 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6885 -1.6928 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1561 -1.1422 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1561 -1.1422 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2662 -1.0125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2662 -1.0125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0248 -0.6174 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0248 -0.6174 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6215 -1.1309 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6215 -1.1309 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1888 -1.6351 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1888 -1.6351 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5534 -1.9250 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5534 -1.9250 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2674 -1.4533 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2674 -1.4533 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.3442 0.2214 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.3442 0.2214 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3851 0.7539 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3851 0.7539 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7283 0.0183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7283 0.0183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0714 0.4123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0714 0.4123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1783 0.3072 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1783 0.3072 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8088 0.8852 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8088 0.8852 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5182 0.5437 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5182 0.5437 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.2197 0.5000 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.2197 0.5000 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6482 0.1387 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6482 0.1387 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3860 -0.0874 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3860 -0.0874 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1366 1.2112 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1366 1.2112 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.3442 1.6163 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.3442 1.6163 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5444 -0.6062 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5444 -0.6062 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3619 -1.6412 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3619 -1.6412 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 3 12 1 0 0 0 0 | + | 3 12 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 18 2 0 0 0 0 | + | 17 18 2 0 0 0 0 |
| − | 18 13 1 0 0 0 0 | + | 18 13 1 0 0 0 0 |
| − | 9 13 1 0 0 0 0 | + | 9 13 1 0 0 0 0 |
| − | 16 19 1 0 0 0 0 | + | 16 19 1 0 0 0 0 |
| − | 1 20 1 0 0 0 0 | + | 1 20 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 2 1 0 0 0 0 | + | 24 2 1 0 0 0 0 |
| − | 15 30 1 0 0 0 0 | + | 15 30 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 34 20 1 0 0 0 0 | + | 34 20 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 36 40 1 0 0 0 0 | + | 36 40 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 26 42 1 0 0 0 0 | + | 26 42 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 40 41 | + | M SAL 1 2 40 41 |
| − | M SBL 1 1 45 | + | M SBL 1 1 45 |
| − | M SMT 1 ^ CH2OH | + | M SMT 1 ^ CH2OH |
| − | M SBV 1 45 0.6184 -0.6675 | + | M SBV 1 45 0.6184 -0.6675 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 42 43 | + | M SAL 2 2 42 43 |
| − | M SBL 2 1 47 | + | M SBL 2 1 47 |
| − | M SMT 2 ^ CH2OH | + | M SMT 2 ^ CH2OH |
| − | M SBV 2 47 0.9230 -0.5247 | + | M SBV 2 47 0.9230 -0.5247 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL3FACDS0006 | + | ID FL3FACDS0006 |
| − | FORMULA C27H30O16 | + | FORMULA C27H30O16 |
| − | EXACTMASS 610.153384912 | + | EXACTMASS 610.153384912 |
| − | AVERAGEMASS 610.5175 | + | AVERAGEMASS 610.5175 |
| − | SMILES C(c(c5O)c(cc(c45)OC(=CC4=O)c(c3)ccc(O)c3O)OC(C2O)OC(C(O)C2O)CO)(C(O)1)OC(C(O)C(O)1)CO | + | SMILES C(c(c5O)c(cc(c45)OC(=CC4=O)c(c3)ccc(O)c3O)OC(C2O)OC(C(O)C2O)CO)(C(O)1)OC(C(O)C(O)1)CO |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
43 47 0 0 0 0 0 0 0 0999 V2000
-0.5959 0.1120 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5959 -0.7063 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1127 -1.1153 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8213 -0.7063 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8213 0.1120 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1127 0.5212 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5299 -1.1153 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2385 -0.7063 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2385 0.1120 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5299 0.5212 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5299 -1.9456 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1127 -1.9333 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1045 0.6524 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8512 0.2212 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5979 0.6524 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5979 1.5146 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8512 1.9456 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1045 1.5146 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.3442 1.9454 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3494 0.5425 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5134 -1.1523 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6885 -1.6928 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1561 -1.1422 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2662 -1.0125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0248 -0.6174 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6215 -1.1309 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1888 -1.6351 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5534 -1.9250 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2674 -1.4533 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.3442 0.2214 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.3851 0.7539 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7283 0.0183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0714 0.4123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1783 0.3072 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8088 0.8852 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5182 0.5437 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.2197 0.5000 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.6482 0.1387 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3860 -0.0874 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1366 1.2112 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.3442 1.6163 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5444 -0.6062 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3619 -1.6412 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
3 12 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 17 1 0 0 0 0
17 18 2 0 0 0 0
18 13 1 0 0 0 0
9 13 1 0 0 0 0
16 19 1 0 0 0 0
1 20 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
24 2 1 0 0 0 0
15 30 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
34 20 1 0 0 0 0
40 41 1 0 0 0 0
36 40 1 0 0 0 0
42 43 1 0 0 0 0
26 42 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 40 41
M SBL 1 1 45
M SMT 1 ^ CH2OH
M SBV 1 45 0.6184 -0.6675
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 42 43
M SBL 2 1 47
M SMT 2 ^ CH2OH
M SBV 2 47 0.9230 -0.5247
S SKP 5
ID FL3FACDS0006
FORMULA C27H30O16
EXACTMASS 610.153384912
AVERAGEMASS 610.5175
SMILES C(c(c5O)c(cc(c45)OC(=CC4=O)c(c3)ccc(O)c3O)OC(C2O)OC(C(O)C2O)CO)(C(O)1)OC(C(O)C(O)1)CO
M END