Mol:FL3FACCS0036
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 44 48 0 0 0 0 0 0 0 0999 V2000 | + | 44 48 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.3017 1.3963 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3017 1.3963 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3017 0.7539 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3017 0.7539 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2546 0.4328 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2546 0.4328 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8109 0.7539 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8109 0.7539 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8109 1.3963 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8109 1.3963 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2546 1.7175 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2546 1.7175 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3672 0.4328 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3672 0.4328 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9235 0.7539 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9235 0.7539 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9235 1.3963 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9235 1.3963 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3672 1.7175 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3672 1.7175 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2546 -0.2094 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2546 -0.2094 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8578 1.7174 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8578 1.7174 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4840 1.7403 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4840 1.7403 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0128 1.4351 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0128 1.4351 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5416 1.7403 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5416 1.7403 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5416 2.3509 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5416 2.3509 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0128 2.6562 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0128 2.6562 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4840 2.3509 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4840 2.3509 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0703 2.6562 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0703 2.6562 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5045 0.7776 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -2.5045 0.7776 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -2.1126 0.1382 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.1126 0.1382 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.5970 0.4476 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.5970 0.4476 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -0.9370 0.3238 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -0.9370 0.3238 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.3907 0.8188 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3907 0.8188 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9476 0.5507 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -1.9476 0.5507 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -3.0927 0.8291 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0927 0.8291 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8476 -0.2044 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8476 -0.2044 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2220 -0.2019 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2220 -0.2019 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0128 3.2667 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0128 3.2667 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2220 -1.0442 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2220 -1.0442 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8554 -1.4109 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8554 -1.4109 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0081 -1.3163 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0081 -1.3163 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3672 -0.2126 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3672 -0.2126 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0474 -2.1025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0474 -2.1025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5173 -2.3699 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5173 -2.3699 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0350 -2.0710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0350 -2.0710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5528 -2.3699 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5528 -2.3699 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5528 -2.9678 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5528 -2.9678 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0350 -3.2667 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0350 -3.2667 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5173 -2.9678 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5173 -2.9678 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0703 -3.2665 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0703 -3.2665 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0696 -2.0715 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0696 -2.0715 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1487 1.4378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1487 1.4378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1038 1.7340 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1038 1.7340 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 3 11 1 0 0 0 0 | + | 3 11 1 0 0 0 0 |
| − | 1 12 1 0 0 0 0 | + | 1 12 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 18 2 0 0 0 0 | + | 17 18 2 0 0 0 0 |
| − | 18 13 1 0 0 0 0 | + | 18 13 1 0 0 0 0 |
| − | 9 13 1 0 0 0 0 | + | 9 13 1 0 0 0 0 |
| − | 16 19 1 0 0 0 0 | + | 16 19 1 0 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 20 1 0 0 0 0 | + | 25 20 1 0 0 0 0 |
| − | 20 26 1 0 0 0 0 | + | 20 26 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 2 1 0 0 0 0 | + | 23 2 1 0 0 0 0 |
| − | 17 29 1 0 0 0 0 | + | 17 29 1 0 0 0 0 |
| − | 28 30 1 0 0 0 0 | + | 28 30 1 0 0 0 0 |
| − | 30 31 2 0 0 0 0 | + | 30 31 2 0 0 0 0 |
| − | 30 32 1 0 0 0 0 | + | 30 32 1 0 0 0 0 |
| − | 7 33 2 0 0 0 0 | + | 7 33 2 0 0 0 0 |
| − | 32 34 2 0 0 0 0 | + | 32 34 2 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 2 0 0 0 0 | + | 35 36 2 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 38 2 0 0 0 0 | + | 37 38 2 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 39 40 2 0 0 0 0 | + | 39 40 2 0 0 0 0 |
| − | 40 35 1 0 0 0 0 | + | 40 35 1 0 0 0 0 |
| − | 38 41 1 0 0 0 0 | + | 38 41 1 0 0 0 0 |
| − | 37 42 1 0 0 0 0 | + | 37 42 1 0 0 0 0 |
| − | 25 43 1 0 0 0 0 | + | 25 43 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 43 44 | + | M SAL 1 2 43 44 |
| − | M SBL 1 1 47 | + | M SBL 1 1 47 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SVB 1 47 -2.1487 1.4378 | + | M SVB 1 47 -2.1487 1.4378 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL3FACCS0036 | + | ID FL3FACCS0036 |
| − | KNApSAcK_ID C00006299 | + | KNApSAcK_ID C00006299 |
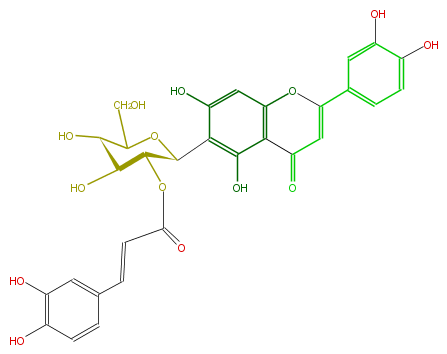
| − | NAME Isoorientin 2''-O-(E)-caffeate;2''-trans-Caffeoylisoorientin | + | NAME Isoorientin 2''-O-(E)-caffeate;2''-trans-Caffeoylisoorientin |
| − | CAS_RN 57186-26-2 | + | CAS_RN 57186-26-2 |
| − | FORMULA C30H26O14 | + | FORMULA C30H26O14 |
| − | EXACTMASS 610.13225554 | + | EXACTMASS 610.13225554 |
| − | AVERAGEMASS 610.51904 | + | AVERAGEMASS 610.51904 |
| − | SMILES O(C(=O)C=Cc(c5)ccc(O)c5O)[C@H]([C@H](c(c4O)c(c(c(c4)3)C(=O)C=C(O3)c(c2)cc(O)c(O)c2)O)1)[C@H]([C@H](C(CO)O1)O)O | + | SMILES O(C(=O)C=Cc(c5)ccc(O)c5O)[C@H]([C@H](c(c4O)c(c(c(c4)3)C(=O)C=C(O3)c(c2)cc(O)c(O)c2)O)1)[C@H]([C@H](C(CO)O1)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
44 48 0 0 0 0 0 0 0 0999 V2000
-0.3017 1.3963 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3017 0.7539 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2546 0.4328 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8109 0.7539 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8109 1.3963 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2546 1.7175 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3672 0.4328 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9235 0.7539 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9235 1.3963 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3672 1.7175 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2546 -0.2094 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8578 1.7174 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4840 1.7403 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0128 1.4351 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5416 1.7403 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5416 2.3509 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0128 2.6562 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4840 2.3509 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0703 2.6562 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5045 0.7776 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-2.1126 0.1382 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.5970 0.4476 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.9370 0.3238 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.3907 0.8188 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9476 0.5507 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-3.0927 0.8291 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8476 -0.2044 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2220 -0.2019 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0128 3.2667 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2220 -1.0442 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8554 -1.4109 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0081 -1.3163 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3672 -0.2126 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0474 -2.1025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5173 -2.3699 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0350 -2.0710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5528 -2.3699 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5528 -2.9678 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0350 -3.2667 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5173 -2.9678 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0703 -3.2665 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0696 -2.0715 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1487 1.4378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1038 1.7340 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
3 11 1 0 0 0 0
1 12 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 17 1 0 0 0 0
17 18 2 0 0 0 0
18 13 1 0 0 0 0
9 13 1 0 0 0 0
16 19 1 0 0 0 0
20 21 1 1 0 0 0
21 22 1 1 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 20 1 0 0 0 0
20 26 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
23 2 1 0 0 0 0
17 29 1 0 0 0 0
28 30 1 0 0 0 0
30 31 2 0 0 0 0
30 32 1 0 0 0 0
7 33 2 0 0 0 0
32 34 2 0 0 0 0
34 35 1 0 0 0 0
35 36 2 0 0 0 0
36 37 1 0 0 0 0
37 38 2 0 0 0 0
38 39 1 0 0 0 0
39 40 2 0 0 0 0
40 35 1 0 0 0 0
38 41 1 0 0 0 0
37 42 1 0 0 0 0
25 43 1 0 0 0 0
43 44 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 43 44
M SBL 1 1 47
M SMT 1 CH2OH
M SVB 1 47 -2.1487 1.4378
S SKP 8
ID FL3FACCS0036
KNApSAcK_ID C00006299
NAME Isoorientin 2''-O-(E)-caffeate;2''-trans-Caffeoylisoorientin
CAS_RN 57186-26-2
FORMULA C30H26O14
EXACTMASS 610.13225554
AVERAGEMASS 610.51904
SMILES O(C(=O)C=Cc(c5)ccc(O)c5O)[C@H]([C@H](c(c4O)c(c(c(c4)3)C(=O)C=C(O3)c(c2)cc(O)c(O)c2)O)1)[C@H]([C@H](C(CO)O1)O)O
M END