Mol:FL3FACCS0029
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 43 47 0 0 0 0 0 0 0 0999 V2000 | + | 43 47 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.4854 0.8888 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4854 0.8888 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4854 0.2464 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4854 0.2464 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0709 -0.0748 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0709 -0.0748 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6272 0.2464 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6272 0.2464 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6272 0.8888 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6272 0.8888 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0709 1.2100 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0709 1.2100 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1835 -0.0748 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1835 -0.0748 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7398 0.2464 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7398 0.2464 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7398 0.8888 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7398 0.8888 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1835 1.2100 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1835 1.2100 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1835 -0.5756 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1835 -0.5756 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0709 -0.7169 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0709 -0.7169 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4196 1.3130 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4196 1.3130 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0058 0.9745 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0058 0.9745 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5920 1.3130 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5920 1.3130 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5920 1.9899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5920 1.9899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0058 2.3283 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0058 2.3283 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4196 1.9899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4196 1.9899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1779 2.3281 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1779 2.3281 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0388 0.0019 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0388 0.0019 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5232 -0.5756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5232 -0.5756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0076 -0.2662 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0076 -0.2662 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3063 -0.3487 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3063 -0.3487 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8013 0.1050 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8013 0.1050 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3582 -0.1631 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3582 -0.1631 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4705 -0.2473 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4705 -0.2473 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1656 -0.6581 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1656 -0.6581 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1619 1.5488 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1619 1.5488 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6982 -0.8021 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6982 -0.8021 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7813 -1.5243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7813 -1.5243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2657 -2.1018 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2657 -2.1018 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7501 -1.7925 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7501 -1.7925 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0488 -1.8750 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0488 -1.8750 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5438 -1.4212 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5438 -1.4212 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1007 -1.6893 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1007 -1.6893 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1779 -1.7533 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1779 -1.7533 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9081 -2.1843 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9081 -2.1843 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4407 -2.3283 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4407 -2.3283 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1779 0.9747 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1779 0.9747 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5200 0.2354 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5200 0.2354 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3169 0.4489 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3169 0.4489 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2389 -1.2909 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2389 -1.2909 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0358 -1.0774 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0358 -1.0774 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 3 12 1 0 0 0 0 | + | 3 12 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 18 2 0 0 0 0 | + | 17 18 2 0 0 0 0 |
| − | 18 13 1 0 0 0 0 | + | 18 13 1 0 0 0 0 |
| − | 9 13 1 0 0 0 0 | + | 9 13 1 0 0 0 0 |
| − | 16 19 1 0 0 0 0 | + | 16 19 1 0 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 20 1 0 0 0 0 | + | 25 20 1 0 0 0 0 |
| − | 20 26 1 0 0 0 0 | + | 20 26 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 2 23 1 0 0 0 0 | + | 2 23 1 0 0 0 0 |
| − | 1 28 1 0 0 0 0 | + | 1 28 1 0 0 0 0 |
| − | 22 29 1 0 0 0 0 | + | 22 29 1 0 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 33 32 1 1 0 0 0 | + | 33 32 1 1 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 30 1 0 0 0 0 | + | 35 30 1 0 0 0 0 |
| − | 30 36 1 0 0 0 0 | + | 30 36 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 29 1 0 0 0 0 | + | 33 29 1 0 0 0 0 |
| − | 15 39 1 0 0 0 0 | + | 15 39 1 0 0 0 0 |
| − | 25 40 1 0 0 0 0 | + | 25 40 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 35 42 1 0 0 0 0 | + | 35 42 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 40 41 | + | M SAL 1 2 40 41 |
| − | M SBL 1 1 44 | + | M SBL 1 1 44 |
| − | M SMT 1 ^CH2OH | + | M SMT 1 ^CH2OH |
| − | M SBV 1 44 -5.5313 7.8872 | + | M SBV 1 44 -5.5313 7.8872 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 42 43 | + | M SAL 2 2 42 43 |
| − | M SBL 2 1 46 | + | M SBL 2 1 46 |
| − | M SMT 2 ^CH2OH | + | M SMT 2 ^CH2OH |
| − | M SBV 2 46 -5.5076 7.8872 | + | M SBV 2 46 -5.5076 7.8872 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL3FACCS0029 | + | ID FL3FACCS0029 |
| − | KNApSAcK_ID C00006210 | + | KNApSAcK_ID C00006210 |
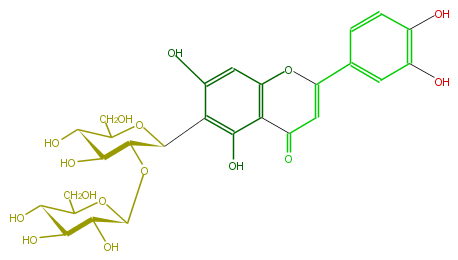
| − | NAME Isoorientin 2''-O-glucopyranoside;Meloside L | + | NAME Isoorientin 2''-O-glucopyranoside;Meloside L |
| − | CAS_RN 55196-48-0 | + | CAS_RN 55196-48-0 |
| − | FORMULA C27H30O16 | + | FORMULA C27H30O16 |
| − | EXACTMASS 610.153384912 | + | EXACTMASS 610.153384912 |
| − | AVERAGEMASS 610.5175 | + | AVERAGEMASS 610.5175 |
| − | SMILES C(C(C1O)OC(OC(C2O)C(c(c3O)c(cc(O4)c(C(C=C(c(c5)ccc(c5O)O)4)=O)3)O)OC(C2O)CO)C(O)C(O)1)O | + | SMILES C(C(C1O)OC(OC(C2O)C(c(c3O)c(cc(O4)c(C(C=C(c(c5)ccc(c5O)O)4)=O)3)O)OC(C2O)CO)C(O)C(O)1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
43 47 0 0 0 0 0 0 0 0999 V2000
-0.4854 0.8888 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4854 0.2464 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0709 -0.0748 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6272 0.2464 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6272 0.8888 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0709 1.2100 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1835 -0.0748 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7398 0.2464 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7398 0.8888 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1835 1.2100 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1835 -0.5756 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0709 -0.7169 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4196 1.3130 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0058 0.9745 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5920 1.3130 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5920 1.9899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0058 2.3283 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4196 1.9899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1779 2.3281 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0388 0.0019 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5232 -0.5756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0076 -0.2662 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3063 -0.3487 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8013 0.1050 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3582 -0.1631 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4705 -0.2473 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1656 -0.6581 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.1619 1.5488 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6982 -0.8021 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7813 -1.5243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2657 -2.1018 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7501 -1.7925 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0488 -1.8750 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5438 -1.4212 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1007 -1.6893 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1779 -1.7533 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9081 -2.1843 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4407 -2.3283 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1779 0.9747 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5200 0.2354 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3169 0.4489 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2389 -1.2909 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0358 -1.0774 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
3 12 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 17 1 0 0 0 0
17 18 2 0 0 0 0
18 13 1 0 0 0 0
9 13 1 0 0 0 0
16 19 1 0 0 0 0
20 21 1 1 0 0 0
21 22 1 1 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 20 1 0 0 0 0
20 26 1 0 0 0 0
21 27 1 0 0 0 0
2 23 1 0 0 0 0
1 28 1 0 0 0 0
22 29 1 0 0 0 0
30 31 1 1 0 0 0
31 32 1 1 0 0 0
33 32 1 1 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 30 1 0 0 0 0
30 36 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 29 1 0 0 0 0
15 39 1 0 0 0 0
25 40 1 0 0 0 0
40 41 1 0 0 0 0
35 42 1 0 0 0 0
42 43 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 40 41
M SBL 1 1 44
M SMT 1 ^CH2OH
M SBV 1 44 -5.5313 7.8872
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 42 43
M SBL 2 1 46
M SMT 2 ^CH2OH
M SBV 2 46 -5.5076 7.8872
S SKP 8
ID FL3FACCS0029
KNApSAcK_ID C00006210
NAME Isoorientin 2''-O-glucopyranoside;Meloside L
CAS_RN 55196-48-0
FORMULA C27H30O16
EXACTMASS 610.153384912
AVERAGEMASS 610.5175
SMILES C(C(C1O)OC(OC(C2O)C(c(c3O)c(cc(O4)c(C(C=C(c(c5)ccc(c5O)O)4)=O)3)O)OC(C2O)CO)C(O)C(O)1)O
M END