Mol:FL3FABDS0001
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 43 47 0 0 0 0 0 0 0 0999 V2000 | + | 43 47 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.2815 0.1067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2815 0.1067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2815 -0.7183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2815 -0.7183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5671 -1.1307 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5671 -1.1307 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1473 -0.7183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1473 -0.7183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1473 0.1067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1473 0.1067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5671 0.5192 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5671 0.5192 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8618 -1.1307 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8618 -1.1307 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5762 -0.7183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5762 -0.7183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5762 0.1067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5762 0.1067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8618 0.5192 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8618 0.5192 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8618 -1.7739 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8618 -1.7739 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5671 -1.9554 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5671 -1.9554 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4492 0.6515 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4492 0.6515 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2022 0.2168 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2022 0.2168 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9550 0.6515 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9550 0.6515 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9550 1.5208 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9550 1.5208 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2022 1.9554 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2022 1.9554 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4492 1.5208 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4492 1.5208 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9957 0.5191 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9957 0.5191 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0462 0.6629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0462 0.6629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3839 -0.0787 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3839 -0.0787 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7218 0.3186 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7218 0.3186 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8212 0.2127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8212 0.2127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4570 0.7955 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4570 0.7955 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1720 0.4510 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1720 0.4510 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.5687 0.3612 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.5687 0.3612 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0271 -0.0787 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0271 -0.0787 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0643 -0.0610 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0643 -0.0610 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1193 -0.6432 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1193 -0.6432 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4571 -1.3849 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4571 -1.3849 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7950 -0.9875 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7950 -0.9875 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8943 -1.0935 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8943 -1.0935 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5300 -0.5107 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5300 -0.5107 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2452 -0.8551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2452 -0.8551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6975 -0.9771 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6975 -0.9771 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1593 -1.3849 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1593 -1.3849 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1545 -1.3573 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1545 -1.3573 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7708 -0.3296 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7708 -0.3296 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7942 -0.0554 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7942 -0.0554 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6015 0.8804 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6015 0.8804 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.6249 1.1546 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.6249 1.1546 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7073 1.9551 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7073 1.9551 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.6249 1.4254 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.6249 1.4254 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 3 12 1 0 0 0 0 | + | 3 12 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 18 2 0 0 0 0 | + | 17 18 2 0 0 0 0 |
| − | 18 13 1 0 0 0 0 | + | 18 13 1 0 0 0 0 |
| − | 9 13 1 0 0 0 0 | + | 9 13 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 20 1 0 0 0 0 | + | 25 20 1 0 0 0 0 |
| − | 20 26 1 0 0 0 0 | + | 20 26 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 19 1 0 0 0 0 | + | 23 19 1 0 0 0 0 |
| − | 29 30 1 1 0 0 0 | + | 29 30 1 1 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 32 31 1 1 0 0 0 | + | 32 31 1 1 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 29 1 0 0 0 0 | + | 34 29 1 0 0 0 0 |
| − | 29 35 1 0 0 0 0 | + | 29 35 1 0 0 0 0 |
| − | 30 36 1 0 0 0 0 | + | 30 36 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 2 1 0 0 0 0 | + | 32 2 1 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 34 38 1 0 0 0 0 | + | 34 38 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 25 40 1 0 0 0 0 | + | 25 40 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 16 42 1 0 0 0 0 | + | 16 42 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 38 39 | + | M SAL 1 2 38 39 |
| − | M SBL 1 1 43 | + | M SBL 1 1 43 |
| − | M SMT 1 ^CH2OH | + | M SMT 1 ^CH2OH |
| − | M SBV 1 43 0.5256 -0.5255 | + | M SBV 1 43 0.5256 -0.5255 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 40 41 | + | M SAL 2 2 40 41 |
| − | M SBL 2 1 45 | + | M SBL 2 1 45 |
| − | M SMT 2 ^CH2OH | + | M SMT 2 ^CH2OH |
| − | M SBV 2 45 0.4294 -0.4294 | + | M SBV 2 45 0.4294 -0.4294 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 42 43 | + | M SAL 3 2 42 43 |
| − | M SBL 3 1 47 | + | M SBL 3 1 47 |
| − | M SMT 3 OCH3 | + | M SMT 3 OCH3 |
| − | M SBV 3 47 -0.7523 -0.4343 | + | M SBV 3 47 -0.7523 -0.4343 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL3FABDS0001 | + | ID FL3FABDS0001 |
| − | FORMULA C28H32O15 | + | FORMULA C28H32O15 |
| − | EXACTMASS 608.174120354 | + | EXACTMASS 608.174120354 |
| − | AVERAGEMASS 608.54468 | + | AVERAGEMASS 608.54468 |
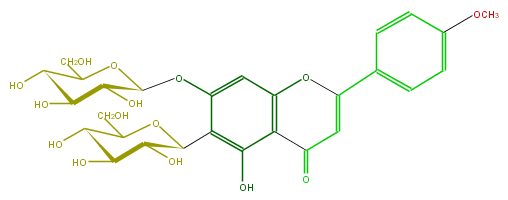
| − | SMILES OC(C(O)1)C(CO)OC(Oc(c5)c(c(c(c53)C(=O)C=C(c(c4)ccc(OC)c4)O3)O)C(C2O)OC(CO)C(O)C2O)C(O)1 | + | SMILES OC(C(O)1)C(CO)OC(Oc(c5)c(c(c(c53)C(=O)C=C(c(c4)ccc(OC)c4)O3)O)C(C2O)OC(CO)C(O)C2O)C(O)1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
43 47 0 0 0 0 0 0 0 0999 V2000
-1.2815 0.1067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2815 -0.7183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5671 -1.1307 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1473 -0.7183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1473 0.1067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5671 0.5192 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8618 -1.1307 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5762 -0.7183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5762 0.1067 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8618 0.5192 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8618 -1.7739 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5671 -1.9554 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4492 0.6515 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2022 0.2168 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9550 0.6515 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9550 1.5208 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2022 1.9554 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4492 1.5208 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9957 0.5191 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.0462 0.6629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3839 -0.0787 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7218 0.3186 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8212 0.2127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4570 0.7955 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1720 0.4510 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.5687 0.3612 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.0271 -0.0787 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0643 -0.0610 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1193 -0.6432 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4571 -1.3849 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7950 -0.9875 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8943 -1.0935 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5300 -0.5107 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2452 -0.8551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.6975 -0.9771 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1593 -1.3849 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1545 -1.3573 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7708 -0.3296 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.7942 -0.0554 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.6015 0.8804 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.6249 1.1546 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.7073 1.9551 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.6249 1.4254 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
3 12 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 17 1 0 0 0 0
17 18 2 0 0 0 0
18 13 1 0 0 0 0
9 13 1 0 0 0 0
1 19 1 0 0 0 0
20 21 1 1 0 0 0
21 22 1 1 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 20 1 0 0 0 0
20 26 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
23 19 1 0 0 0 0
29 30 1 1 0 0 0
30 31 1 1 0 0 0
32 31 1 1 0 0 0
32 33 1 0 0 0 0
33 34 1 0 0 0 0
34 29 1 0 0 0 0
29 35 1 0 0 0 0
30 36 1 0 0 0 0
31 37 1 0 0 0 0
32 2 1 0 0 0 0
38 39 1 0 0 0 0
34 38 1 0 0 0 0
40 41 1 0 0 0 0
25 40 1 0 0 0 0
42 43 1 0 0 0 0
16 42 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 38 39
M SBL 1 1 43
M SMT 1 ^CH2OH
M SBV 1 43 0.5256 -0.5255
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 40 41
M SBL 2 1 45
M SMT 2 ^CH2OH
M SBV 2 45 0.4294 -0.4294
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 42 43
M SBL 3 1 47
M SMT 3 OCH3
M SBV 3 47 -0.7523 -0.4343
S SKP 5
ID FL3FABDS0001
FORMULA C28H32O15
EXACTMASS 608.174120354
AVERAGEMASS 608.54468
SMILES OC(C(O)1)C(CO)OC(Oc(c5)c(c(c(c53)C(=O)C=C(c(c4)ccc(OC)c4)O3)O)C(C2O)OC(CO)C(O)C2O)C(O)1
M END