Mol:FL3FAAGS0064
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 42 46 0 0 0 0 0 0 0 0999 V2000 | + | 42 46 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.9176 1.7526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9176 1.7526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9176 0.9276 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9176 0.9276 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2032 0.5151 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2032 0.5151 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5113 0.9276 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5113 0.9276 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5113 1.7526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5113 1.7526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2032 2.1651 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2032 2.1651 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2258 0.5151 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2258 0.5151 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9403 0.9276 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9403 0.9276 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9403 1.7526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9403 1.7526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2258 2.1651 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2258 2.1651 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6547 2.1651 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6547 2.1651 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3692 1.7526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3692 1.7526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0837 2.1651 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0837 2.1651 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0837 2.9901 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0837 2.9901 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3692 3.4026 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3692 3.4026 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6547 2.9901 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6547 2.9901 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2258 -0.3099 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2258 -0.3099 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2032 -0.3099 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2032 -0.3099 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8300 3.4210 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8300 3.4210 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6372 2.0996 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6372 2.0996 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3467 2.1764 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3467 2.1764 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9341 1.4617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9341 1.4617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1358 1.6709 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1358 1.6709 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3423 1.4440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3423 1.4440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7549 2.1587 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7549 2.1587 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5533 1.9497 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5533 1.9497 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7102 2.5355 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7102 2.5355 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1901 2.8125 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1901 2.8125 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8300 1.6932 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8300 1.6932 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3828 1.7207 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3828 1.7207 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3667 0.8868 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3667 0.8868 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7858 0.1775 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7858 0.1775 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6097 0.1859 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6097 0.1859 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3811 -0.5401 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3811 -0.5401 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5573 -0.5485 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5573 -0.5485 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1526 -1.2661 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1526 -1.2661 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5723 -1.9764 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5723 -1.9764 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1671 -2.6950 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1671 -2.6950 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3421 -2.7033 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3421 -2.7033 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9224 -1.9931 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9224 -1.9931 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3276 -1.2745 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3276 -1.2745 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9374 -3.4210 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9374 -3.4210 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 9 11 1 0 0 0 0 | + | 9 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 7 17 2 0 0 0 0 | + | 7 17 2 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 14 19 1 0 0 0 0 | + | 14 19 1 0 0 0 0 |
| − | 20 1 1 0 0 0 0 | + | 20 1 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 21 29 1 0 0 0 0 | + | 21 29 1 0 0 0 0 |
| − | 22 30 1 0 0 0 0 | + | 22 30 1 0 0 0 0 |
| − | 24 20 1 0 0 0 0 | + | 24 20 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 2 0 0 0 0 | + | 32 33 2 0 0 0 0 |
| − | 32 34 1 0 0 0 0 | + | 32 34 1 0 0 0 0 |
| − | 34 35 2 0 0 0 0 | + | 34 35 2 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 2 0 0 0 0 | + | 36 37 2 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 2 0 0 0 0 | + | 38 39 2 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 40 41 2 0 0 0 0 | + | 40 41 2 0 0 0 0 |
| − | 41 36 1 0 0 0 0 | + | 41 36 1 0 0 0 0 |
| − | 39 42 1 0 0 0 0 | + | 39 42 1 0 0 0 0 |
| − | 31 23 1 0 0 0 0 | + | 31 23 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL3FAAGS0064 | + | ID FL3FAAGS0064 |
| − | KNApSAcK_ID C00013614 | + | KNApSAcK_ID C00013614 |
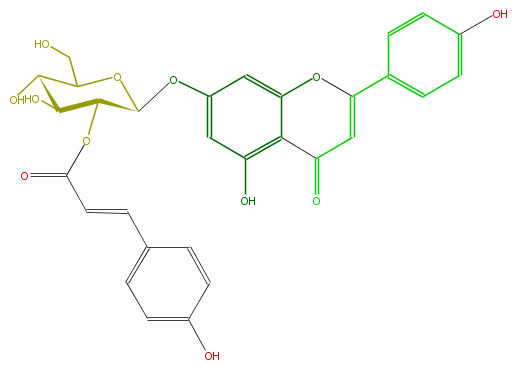
| − | NAME Apigenin 7-(2''-E-p-coumaroylglucoside;Echitin (glycoside);(E)-5-Hydroxy-2-(4-hydroxyphenyl)-7-[[2-O-[3-(4-hydroxyphenyl)-1-oxo-2-propenyl]-beta-D-glucopyranosyl]oxy]-4H-1-benzopyran-4-one | + | NAME Apigenin 7-(2''-E-p-coumaroylglucoside;Echitin (glycoside);(E)-5-Hydroxy-2-(4-hydroxyphenyl)-7-[[2-O-[3-(4-hydroxyphenyl)-1-oxo-2-propenyl]-beta-D-glucopyranosyl]oxy]-4H-1-benzopyran-4-one |
| − | CAS_RN 177535-49-8 | + | CAS_RN 177535-49-8 |
| − | FORMULA C30H26O12 | + | FORMULA C30H26O12 |
| − | EXACTMASS 578.1424262959999 | + | EXACTMASS 578.1424262959999 |
| − | AVERAGEMASS 578.5202400000001 | + | AVERAGEMASS 578.5202400000001 |
| − | SMILES O(C5CO)C(C(C(C5O)O)OC(=O)C=Cc(c4)ccc(c4)O)Oc(c3)cc(O1)c(c(O)3)C(C=C(c(c2)ccc(O)c2)1)=O | + | SMILES O(C5CO)C(C(C(C5O)O)OC(=O)C=Cc(c4)ccc(c4)O)Oc(c3)cc(O1)c(c(O)3)C(C=C(c(c2)ccc(O)c2)1)=O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
42 46 0 0 0 0 0 0 0 0999 V2000
-0.9176 1.7526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9176 0.9276 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2032 0.5151 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5113 0.9276 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5113 1.7526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2032 2.1651 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2258 0.5151 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9403 0.9276 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9403 1.7526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2258 2.1651 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6547 2.1651 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3692 1.7526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0837 2.1651 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0837 2.9901 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3692 3.4026 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6547 2.9901 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2258 -0.3099 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2032 -0.3099 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.8300 3.4210 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6372 2.0996 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.3467 2.1764 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9341 1.4617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1358 1.6709 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3423 1.4440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7549 2.1587 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5533 1.9497 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7102 2.5355 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1901 2.8125 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.8300 1.6932 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.3828 1.7207 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3667 0.8868 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7858 0.1775 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.6097 0.1859 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3811 -0.5401 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5573 -0.5485 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1526 -1.2661 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5723 -1.9764 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1671 -2.6950 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3421 -2.7033 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9224 -1.9931 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3276 -1.2745 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9374 -3.4210 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
7 17 2 0 0 0 0
3 18 1 0 0 0 0
14 19 1 0 0 0 0
20 1 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
26 27 1 0 0 0 0
27 28 1 0 0 0 0
21 29 1 0 0 0 0
22 30 1 0 0 0 0
24 20 1 0 0 0 0
31 32 1 0 0 0 0
32 33 2 0 0 0 0
32 34 1 0 0 0 0
34 35 2 0 0 0 0
35 36 1 0 0 0 0
36 37 2 0 0 0 0
37 38 1 0 0 0 0
38 39 2 0 0 0 0
39 40 1 0 0 0 0
40 41 2 0 0 0 0
41 36 1 0 0 0 0
39 42 1 0 0 0 0
31 23 1 0 0 0 0
S SKP 8
ID FL3FAAGS0064
KNApSAcK_ID C00013614
NAME Apigenin 7-(2''-E-p-coumaroylglucoside;Echitin (glycoside);(E)-5-Hydroxy-2-(4-hydroxyphenyl)-7-[[2-O-[3-(4-hydroxyphenyl)-1-oxo-2-propenyl]-beta-D-glucopyranosyl]oxy]-4H-1-benzopyran-4-one
CAS_RN 177535-49-8
FORMULA C30H26O12
EXACTMASS 578.1424262959999
AVERAGEMASS 578.5202400000001
SMILES O(C5CO)C(C(C(C5O)O)OC(=O)C=Cc(c4)ccc(c4)O)Oc(c3)cc(O1)c(c(O)3)C(C=C(c(c2)ccc(O)c2)1)=O
M END