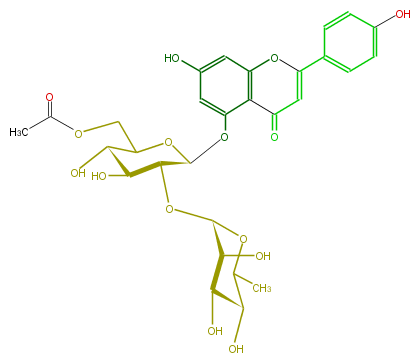
Mol:FL3FAAGS0051
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 44 48 0 0 0 0 0 0 0 0999 V2000 | + | 44 48 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.2471 2.1836 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2471 2.1836 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2471 1.6107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2471 1.6107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2492 1.3242 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2492 1.3242 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7454 1.6107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7454 1.6107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7454 2.1836 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7454 2.1836 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2492 2.4701 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2492 2.4701 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2416 1.3242 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2416 1.3242 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7378 1.6107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7378 1.6107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7378 2.1836 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7378 2.1836 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2416 2.4701 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2416 2.4701 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2416 0.8774 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2416 0.8774 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2339 2.4700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2339 2.4700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7396 2.1780 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7396 2.1780 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2453 2.4700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2453 2.4700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2453 3.0540 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2453 3.0540 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7396 3.3460 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7396 3.3460 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2339 3.0540 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2339 3.0540 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7509 3.3459 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7509 3.3459 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2492 0.8771 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2492 0.8771 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7575 0.1736 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7575 0.1736 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0967 0.6670 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0967 0.6670 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6651 0.0973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6651 0.0973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0437 0.3390 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0437 0.3390 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4440 0.3454 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4440 0.3454 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8798 0.7813 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8798 0.7813 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5146 0.5533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5146 0.5533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1884 0.0973 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1884 0.0973 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8600 -0.5451 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8600 -0.5451 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8714 1.1714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8714 1.1714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6792 0.9550 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6792 0.9550 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2457 1.2820 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2457 1.2820 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7509 0.9903 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7509 0.9903 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2457 1.6825 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2457 1.6825 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0061 -2.1957 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0061 -2.1957 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6221 -2.5584 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6221 -2.5584 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4227 -1.8609 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4227 -1.8609 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6221 -1.1592 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6221 -1.1592 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0061 -0.7964 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0061 -0.7964 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1931 -1.4940 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1931 -1.4940 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8847 -2.1277 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8847 -2.1277 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9552 -1.4940 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9552 -1.4940 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4111 -3.3460 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4111 -3.3460 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0061 -3.0025 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0061 -3.0025 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7219 2.4578 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7219 2.4578 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 15 18 1 0 0 0 0 | + | 15 18 1 0 0 0 0 |
| − | 19 3 1 0 0 0 0 | + | 19 3 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 22 27 1 0 0 0 0 | + | 22 27 1 0 0 0 0 |
| − | 23 28 1 0 0 0 0 | + | 23 28 1 0 0 0 0 |
| − | 20 21 1 0 0 0 0 | + | 20 21 1 0 0 0 0 |
| − | 26 29 1 0 0 0 0 | + | 26 29 1 0 0 0 0 |
| − | 29 30 1 0 0 0 0 | + | 29 30 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 31 33 2 0 0 0 0 | + | 31 33 2 0 0 0 0 |
| − | 35 34 1 1 0 0 0 | + | 35 34 1 1 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 1 1 0 0 0 | + | 38 39 1 1 0 0 0 |
| − | 39 34 1 1 0 0 0 | + | 39 34 1 1 0 0 0 |
| − | 36 40 1 0 0 0 0 | + | 36 40 1 0 0 0 0 |
| − | 28 38 1 0 0 0 0 | + | 28 38 1 0 0 0 0 |
| − | 39 41 1 0 0 0 0 | + | 39 41 1 0 0 0 0 |
| − | 35 42 1 0 0 0 0 | + | 35 42 1 0 0 0 0 |
| − | 34 43 1 0 0 0 0 | + | 34 43 1 0 0 0 0 |
| − | 24 19 1 0 0 0 0 | + | 24 19 1 0 0 0 0 |
| − | 1 44 1 0 0 0 0 | + | 1 44 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL3FAAGS0051 | + | ID FL3FAAGS0051 |
| − | KNApSAcK_ID C00004191 | + | KNApSAcK_ID C00004191 |
| − | NAME Apigenin 5-rhamnosyl-(1->2)-(6''-acetylglucoside) | + | NAME Apigenin 5-rhamnosyl-(1->2)-(6''-acetylglucoside) |
| − | CAS_RN 125300-52-9 | + | CAS_RN 125300-52-9 |
| − | FORMULA C29H32O15 | + | FORMULA C29H32O15 |
| − | EXACTMASS 620.174120354 | + | EXACTMASS 620.174120354 |
| − | AVERAGEMASS 620.55538 | + | AVERAGEMASS 620.55538 |
| − | SMILES C(O)(C(O)1)C(OC(Oc(c5)c(C3=O)c(cc(O)5)OC(c(c4)ccc(c4)O)=C3)C1OC(C2O)OC(C)C(C2O)O)COC(C)=O | + | SMILES C(O)(C(O)1)C(OC(Oc(c5)c(C3=O)c(cc(O)5)OC(c(c4)ccc(c4)O)=C3)C1OC(C2O)OC(C)C(C2O)O)COC(C)=O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
44 48 0 0 0 0 0 0 0 0999 V2000
-0.2471 2.1836 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2471 1.6107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2492 1.3242 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7454 1.6107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7454 2.1836 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2492 2.4701 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2416 1.3242 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7378 1.6107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7378 2.1836 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2416 2.4701 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2416 0.8774 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2339 2.4700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7396 2.1780 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2453 2.4700 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2453 3.0540 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7396 3.3460 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2339 3.0540 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7509 3.3459 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2492 0.8771 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7575 0.1736 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0967 0.6670 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6651 0.0973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0437 0.3390 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4440 0.3454 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8798 0.7813 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.5146 0.5533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1884 0.0973 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8600 -0.5451 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8714 1.1714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6792 0.9550 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2457 1.2820 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7509 0.9903 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2457 1.6825 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0061 -2.1957 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6221 -2.5584 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4227 -1.8609 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6221 -1.1592 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0061 -0.7964 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1931 -1.4940 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8847 -2.1277 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9552 -1.4940 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4111 -3.3460 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0061 -3.0025 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7219 2.4578 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
15 18 1 0 0 0 0
19 3 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
22 27 1 0 0 0 0
23 28 1 0 0 0 0
20 21 1 0 0 0 0
26 29 1 0 0 0 0
29 30 1 0 0 0 0
30 31 1 0 0 0 0
31 32 1 0 0 0 0
31 33 2 0 0 0 0
35 34 1 1 0 0 0
35 36 1 0 0 0 0
36 37 1 0 0 0 0
37 38 1 0 0 0 0
38 39 1 1 0 0 0
39 34 1 1 0 0 0
36 40 1 0 0 0 0
28 38 1 0 0 0 0
39 41 1 0 0 0 0
35 42 1 0 0 0 0
34 43 1 0 0 0 0
24 19 1 0 0 0 0
1 44 1 0 0 0 0
S SKP 8
ID FL3FAAGS0051
KNApSAcK_ID C00004191
NAME Apigenin 5-rhamnosyl-(1->2)-(6''-acetylglucoside)
CAS_RN 125300-52-9
FORMULA C29H32O15
EXACTMASS 620.174120354
AVERAGEMASS 620.55538
SMILES C(O)(C(O)1)C(OC(Oc(c5)c(C3=O)c(cc(O)5)OC(c(c4)ccc(c4)O)=C3)C1OC(C2O)OC(C)C(C2O)O)COC(C)=O
M END