Mol:FL3FAAGS0047
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 53 58 0 0 0 0 0 0 0 0999 V2000 | + | 53 58 0 0 0 0 0 0 0 0999 V2000 |
| − | 0.5320 0.8905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5320 0.8905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5320 0.3176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5320 0.3176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0282 0.0311 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0282 0.0311 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5244 0.3176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5244 0.3176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5244 0.8905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5244 0.8905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0282 1.1770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0282 1.1770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0206 0.0311 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0206 0.0311 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5169 0.3176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5169 0.3176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5169 0.8905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5169 0.8905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0206 1.1770 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0206 1.1770 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2095 -0.3737 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2095 -0.3737 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0129 1.1769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0129 1.1769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5186 0.8849 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5186 0.8849 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0244 1.1769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0244 1.1769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0244 1.7609 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0244 1.7609 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5186 2.0529 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5186 2.0529 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0129 1.7609 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0129 1.7609 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0359 1.1769 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0359 1.1769 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5300 2.0528 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5300 2.0528 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9507 -0.4091 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9507 -0.4091 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2476 0.8935 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -2.2476 0.8935 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -1.8599 0.3817 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.8599 0.3817 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.3016 0.5988 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.3016 0.5988 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -0.6892 0.4942 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -0.6892 0.4942 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.1543 0.9962 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1543 0.9962 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7247 0.7914 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -1.7247 0.7914 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -3.0401 0.5962 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0401 0.5962 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4643 0.3523 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4643 0.3523 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9817 0.0618 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9817 0.0618 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6426 1.1882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6426 1.1882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2854 1.8243 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2854 1.8243 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8234 2.2863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8234 2.2863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0904 2.7488 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0904 2.7488 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2909 2.2863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2909 2.2863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2529 2.7200 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2529 2.7200 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7825 2.9394 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7825 2.9394 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3067 2.6062 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3067 2.6062 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2197 2.8517 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2197 2.8517 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2703 3.4303 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2703 3.4303 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2055 3.7634 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2055 3.7634 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7318 3.5180 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7318 3.5180 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7961 3.6755 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7961 3.6755 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0456 -1.4883 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0456 -1.4883 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5300 -1.7680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5300 -1.7680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5300 -2.3274 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5300 -2.3274 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0456 -2.6070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0456 -2.6070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5612 -2.3274 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5612 -2.3274 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5612 -1.7680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5612 -1.7680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0456 -3.1663 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0456 -3.1663 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5031 0.1332 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5031 0.1332 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0856 0.4695 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0856 0.4695 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5031 -0.4783 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5031 -0.4783 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0190 -0.7762 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0190 -0.7762 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 15 19 1 0 0 0 0 | + | 15 19 1 0 0 0 0 |
| − | 20 3 1 0 0 0 0 | + | 20 3 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 18 24 1 0 0 0 0 | + | 18 24 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 2 0 0 0 0 | + | 32 33 2 0 0 0 0 |
| − | 32 34 1 0 0 0 0 | + | 32 34 1 0 0 0 0 |
| − | 34 35 2 0 0 0 0 | + | 34 35 2 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 2 0 0 0 0 | + | 36 37 2 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 2 0 0 0 0 | + | 38 39 2 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 40 41 2 0 0 0 0 | + | 40 41 2 0 0 0 0 |
| − | 41 36 1 0 0 0 0 | + | 41 36 1 0 0 0 0 |
| − | 39 42 1 0 0 0 0 | + | 39 42 1 0 0 0 0 |
| − | 43 44 2 0 0 0 0 | + | 43 44 2 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 45 46 2 0 0 0 0 | + | 45 46 2 0 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 47 48 2 0 0 0 0 | + | 47 48 2 0 0 0 0 |
| − | 48 43 1 0 0 0 0 | + | 48 43 1 0 0 0 0 |
| − | 46 49 1 0 0 0 0 | + | 46 49 1 0 0 0 0 |
| − | 27 50 1 0 0 0 0 | + | 27 50 1 0 0 0 0 |
| − | 50 51 2 0 0 0 0 | + | 50 51 2 0 0 0 0 |
| − | 50 52 1 0 0 0 0 | + | 50 52 1 0 0 0 0 |
| − | 52 53 2 0 0 0 0 | + | 52 53 2 0 0 0 0 |
| − | 53 43 1 0 0 0 0 | + | 53 43 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL3FAAGS0047 | + | ID FL3FAAGS0047 |
| − | KNApSAcK_ID C00004187 | + | KNApSAcK_ID C00004187 |
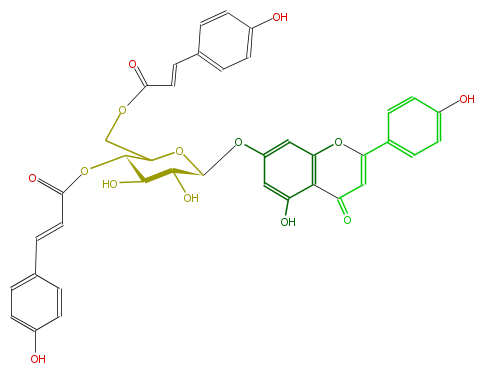
| − | NAME Apigenin 7-(4'',6''-di-p-coumarylglucoside) | + | NAME Apigenin 7-(4'',6''-di-p-coumarylglucoside) |
| − | CAS_RN 87562-09-2 | + | CAS_RN 87562-09-2 |
| − | FORMULA C39H32O14 | + | FORMULA C39H32O14 |
| − | EXACTMASS 724.179205732 | + | EXACTMASS 724.179205732 |
| − | AVERAGEMASS 724.6629800000001 | + | AVERAGEMASS 724.6629800000001 |
| − | SMILES c(c6)c(ccc(O)6)C=CC(=O)O[C@@H](C4COC(C=Cc(c5)ccc(O)c5)=O)[C@@H]([C@@H]([C@@H](O4)Oc(c1)cc(c(C(=O)3)c(OC(=C3)c(c2)ccc(c2)O)1)O)O)O | + | SMILES c(c6)c(ccc(O)6)C=CC(=O)O[C@@H](C4COC(C=Cc(c5)ccc(O)c5)=O)[C@@H]([C@@H]([C@@H](O4)Oc(c1)cc(c(C(=O)3)c(OC(=C3)c(c2)ccc(c2)O)1)O)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
53 58 0 0 0 0 0 0 0 0999 V2000
0.5320 0.8905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5320 0.3176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0282 0.0311 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5244 0.3176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5244 0.8905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0282 1.1770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0206 0.0311 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5169 0.3176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5169 0.8905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0206 1.1770 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2095 -0.3737 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0129 1.1769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5186 0.8849 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0244 1.1769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0244 1.7609 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5186 2.0529 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0129 1.7609 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0359 1.1769 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.5300 2.0528 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9507 -0.4091 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2476 0.8935 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-1.8599 0.3817 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.3016 0.5988 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.6892 0.4942 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.1543 0.9962 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7247 0.7914 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-3.0401 0.5962 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4643 0.3523 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9817 0.0618 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6426 1.1882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2854 1.8243 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8234 2.2863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0904 2.7488 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2909 2.2863 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2529 2.7200 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7825 2.9394 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3067 2.6062 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2197 2.8517 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2703 3.4303 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2055 3.7634 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7318 3.5180 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7961 3.6755 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0456 -1.4883 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.5300 -1.7680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.5300 -2.3274 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0456 -2.6070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5612 -2.3274 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5612 -1.7680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0456 -3.1663 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5031 0.1332 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0856 0.4695 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5031 -0.4783 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0190 -0.7762 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
15 19 1 0 0 0 0
20 3 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
26 30 1 0 0 0 0
18 24 1 0 0 0 0
30 31 1 0 0 0 0
31 32 1 0 0 0 0
32 33 2 0 0 0 0
32 34 1 0 0 0 0
34 35 2 0 0 0 0
35 36 1 0 0 0 0
36 37 2 0 0 0 0
37 38 1 0 0 0 0
38 39 2 0 0 0 0
39 40 1 0 0 0 0
40 41 2 0 0 0 0
41 36 1 0 0 0 0
39 42 1 0 0 0 0
43 44 2 0 0 0 0
44 45 1 0 0 0 0
45 46 2 0 0 0 0
46 47 1 0 0 0 0
47 48 2 0 0 0 0
48 43 1 0 0 0 0
46 49 1 0 0 0 0
27 50 1 0 0 0 0
50 51 2 0 0 0 0
50 52 1 0 0 0 0
52 53 2 0 0 0 0
53 43 1 0 0 0 0
S SKP 8
ID FL3FAAGS0047
KNApSAcK_ID C00004187
NAME Apigenin 7-(4'',6''-di-p-coumarylglucoside)
CAS_RN 87562-09-2
FORMULA C39H32O14
EXACTMASS 724.179205732
AVERAGEMASS 724.6629800000001
SMILES c(c6)c(ccc(O)6)C=CC(=O)O[C@@H](C4COC(C=Cc(c5)ccc(O)c5)=O)[C@@H]([C@@H]([C@@H](O4)Oc(c1)cc(c(C(=O)3)c(OC(=C3)c(c2)ccc(c2)O)1)O)O)O
M END