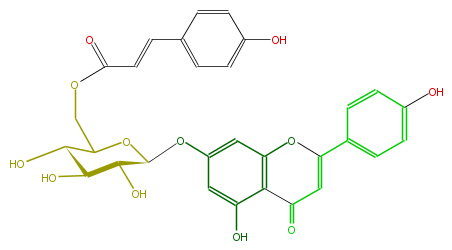
Mol:FL3FAAGS0036
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 42 46 0 0 0 0 0 0 0 0999 V2000 | + | 42 46 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.3625 -0.6387 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3625 -0.6387 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3625 -1.2810 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3625 -1.2810 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1938 -1.6022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1938 -1.6022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7501 -1.2810 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7501 -1.2810 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7501 -0.6387 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7501 -0.6387 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1938 -0.3175 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1938 -0.3175 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3064 -1.6022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3064 -1.6022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8627 -1.2810 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8627 -1.2810 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8627 -0.6387 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8627 -0.6387 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3064 -0.3175 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3064 -0.3175 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3064 -2.1030 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3064 -2.1030 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4188 -0.3176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4188 -0.3176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9858 -0.6449 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9858 -0.6449 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5527 -0.3176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5527 -0.3176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5527 0.3371 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5527 0.3371 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9858 0.6644 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9858 0.6644 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4188 0.3371 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4188 0.3371 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9186 -0.3176 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9186 -0.3176 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1938 -2.2443 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1938 -2.2443 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1196 0.6643 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1196 0.6643 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2312 -0.4497 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2312 -0.4497 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7965 -1.0235 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7965 -1.0235 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1706 -0.7801 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1706 -0.7801 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5666 -0.7736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5666 -0.7736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0055 -0.3346 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0055 -0.3346 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6449 -0.5642 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6449 -0.5642 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1196 -0.7830 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1196 -0.7830 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4741 -1.0565 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4741 -1.0565 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8119 -1.3821 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8119 -1.3821 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0348 0.1112 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0348 0.1112 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0348 0.8057 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0348 0.8057 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4213 1.1599 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4213 1.1599 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7350 1.7031 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7350 1.7031 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8290 1.1599 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8290 1.1599 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5163 1.7016 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5163 1.7016 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9020 1.7016 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9020 1.7016 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5886 1.1589 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5886 1.1589 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0381 1.1589 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0381 1.1589 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3514 1.7016 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3514 1.7016 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0381 2.2443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0381 2.2443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5886 2.2443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5886 2.2443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9771 1.7016 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9771 1.7016 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 18 1 0 0 0 0 | + | 24 18 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 2 0 0 0 0 | + | 32 33 2 0 0 0 0 |
| − | 32 34 1 0 0 0 0 | + | 32 34 1 0 0 0 0 |
| − | 34 35 2 0 0 0 0 | + | 34 35 2 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 2 0 0 0 0 | + | 36 37 2 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 2 0 0 0 0 | + | 38 39 2 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 40 41 2 0 0 0 0 | + | 40 41 2 0 0 0 0 |
| − | 41 36 1 0 0 0 0 | + | 41 36 1 0 0 0 0 |
| − | 39 42 1 0 0 0 0 | + | 39 42 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL3FAAGS0036 | + | ID FL3FAAGS0036 |
| − | KNApSAcK_ID C00004172 | + | KNApSAcK_ID C00004172 |
| − | NAME Apigenin 7-(6''-p-coumarylglucoside) | + | NAME Apigenin 7-(6''-p-coumarylglucoside) |
| − | CAS_RN 105815-90-5 | + | CAS_RN 105815-90-5 |
| − | FORMULA C30H26O12 | + | FORMULA C30H26O12 |
| − | EXACTMASS 578.1424262959999 | + | EXACTMASS 578.1424262959999 |
| − | AVERAGEMASS 578.5202400000001 | + | AVERAGEMASS 578.5202400000001 |
| − | SMILES c(O)(c5)ccc(c5)C(O4)=CC(c(c34)c(cc(c3)OC(O1)C(C(O)C(C1COC(C=Cc(c2)ccc(O)c2)=O)O)O)O)=O | + | SMILES c(O)(c5)ccc(c5)C(O4)=CC(c(c34)c(cc(c3)OC(O1)C(C(O)C(C1COC(C=Cc(c2)ccc(O)c2)=O)O)O)O)=O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
42 46 0 0 0 0 0 0 0 0999 V2000
-0.3625 -0.6387 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3625 -1.2810 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1938 -1.6022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7501 -1.2810 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7501 -0.6387 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1938 -0.3175 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3064 -1.6022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8627 -1.2810 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8627 -0.6387 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3064 -0.3175 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3064 -2.1030 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4188 -0.3176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9858 -0.6449 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5527 -0.3176 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5527 0.3371 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9858 0.6644 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4188 0.3371 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9186 -0.3176 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1938 -2.2443 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1196 0.6643 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2312 -0.4497 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7965 -1.0235 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1706 -0.7801 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5666 -0.7736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0055 -0.3346 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6449 -0.5642 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1196 -0.7830 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4741 -1.0565 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8119 -1.3821 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0348 0.1112 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0348 0.8057 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4213 1.1599 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7350 1.7031 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8290 1.1599 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5163 1.7016 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9020 1.7016 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5886 1.1589 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0381 1.1589 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3514 1.7016 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0381 2.2443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5886 2.2443 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9771 1.7016 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
3 19 1 0 0 0 0
15 20 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
24 18 1 0 0 0 0
26 30 1 0 0 0 0
30 31 1 0 0 0 0
31 32 1 0 0 0 0
32 33 2 0 0 0 0
32 34 1 0 0 0 0
34 35 2 0 0 0 0
35 36 1 0 0 0 0
36 37 2 0 0 0 0
37 38 1 0 0 0 0
38 39 2 0 0 0 0
39 40 1 0 0 0 0
40 41 2 0 0 0 0
41 36 1 0 0 0 0
39 42 1 0 0 0 0
S SKP 8
ID FL3FAAGS0036
KNApSAcK_ID C00004172
NAME Apigenin 7-(6''-p-coumarylglucoside)
CAS_RN 105815-90-5
FORMULA C30H26O12
EXACTMASS 578.1424262959999
AVERAGEMASS 578.5202400000001
SMILES c(O)(c5)ccc(c5)C(O4)=CC(c(c34)c(cc(c3)OC(O1)C(C(O)C(C1COC(C=Cc(c2)ccc(O)c2)=O)O)O)O)=O
M END