Mol:FL3FAAGS0030
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 52 57 0 0 0 0 0 0 0 0999 V2000 | + | 52 57 0 0 0 0 0 0 0 0999 V2000 |
| − | 0.0049 -1.6485 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0049 -1.6485 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0049 -2.4581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0049 -2.4581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7060 -2.8630 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7060 -2.8630 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4071 -2.4581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4071 -2.4581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4071 -1.6485 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4071 -1.6485 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7060 -1.2437 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7060 -1.2437 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1082 -2.8630 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1082 -2.8630 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8094 -2.4581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8094 -2.4581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8094 -1.6485 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8094 -1.6485 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1082 -1.2437 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1082 -1.2437 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1082 -3.4942 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1082 -3.4942 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5103 -1.2439 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5103 -1.2439 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2249 -1.6565 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2249 -1.6565 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9393 -1.2439 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9393 -1.2439 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9393 -0.4188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9393 -0.4188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2249 -0.0062 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2249 -0.0062 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5103 -0.4188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5103 -0.4188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6328 -1.2453 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6328 -1.2453 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7060 -3.6723 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7060 -3.6723 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.6539 -0.0063 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.6539 -0.0063 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9859 1.2063 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9859 1.2063 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.7777 0.7491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.7777 0.7491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.5265 1.6282 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.5265 1.6282 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.7777 2.5126 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.7777 2.5126 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9859 2.9698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9859 2.9698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.2372 2.0908 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.2372 2.0908 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1742 3.6723 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1742 3.6723 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.7777 3.1695 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.7777 3.1695 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.9921 1.2864 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.9921 1.2864 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6935 -1.0739 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6935 -1.0739 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0437 -1.9317 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0437 -1.9317 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1079 -1.5678 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1079 -1.5678 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2048 -1.5581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2048 -1.5581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8609 -0.9018 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8609 -0.9018 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6797 -1.3338 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6797 -1.3338 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0574 -0.7724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0574 -0.7724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2420 -1.2928 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2420 -1.2928 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7027 -1.8919 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7027 -1.8919 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4540 -1.9452 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4540 -1.9452 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.3509 -0.4940 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.3509 -0.4940 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5591 -0.9512 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5591 -0.9512 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8103 -0.0722 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8103 -0.0722 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5591 0.8124 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5591 0.8124 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.3509 1.2696 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.3509 1.2696 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0996 0.3904 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0996 0.3904 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8137 -0.6321 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8137 -0.6321 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0879 1.8683 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0879 1.8683 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5591 1.4776 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5591 1.4776 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.6874 0.7926 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.6874 0.7926 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.9921 -0.5573 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.9921 -0.5573 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5898 2.5526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5898 2.5526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6290 1.7367 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6290 1.7367 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 22 21 1 1 0 0 0 | + | 22 21 1 1 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 1 0 0 0 | + | 25 26 1 1 0 0 0 |
| − | 26 21 1 1 0 0 0 | + | 26 21 1 1 0 0 0 |
| − | 25 27 1 0 0 0 0 | + | 25 27 1 0 0 0 0 |
| − | 24 28 1 0 0 0 0 | + | 24 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 22 20 1 0 0 0 0 | + | 22 20 1 0 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 33 32 1 1 0 0 0 | + | 33 32 1 1 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 30 1 0 0 0 0 | + | 35 30 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 30 37 1 0 0 0 0 | + | 30 37 1 0 0 0 0 |
| − | 31 38 1 0 0 0 0 | + | 31 38 1 0 0 0 0 |
| − | 32 39 1 0 0 0 0 | + | 32 39 1 0 0 0 0 |
| − | 41 40 1 1 0 0 0 | + | 41 40 1 1 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 44 45 1 1 0 0 0 | + | 44 45 1 1 0 0 0 |
| − | 45 40 1 1 0 0 0 | + | 45 40 1 1 0 0 0 |
| − | 41 46 1 0 0 0 0 | + | 41 46 1 0 0 0 0 |
| − | 46 36 1 0 0 0 0 | + | 46 36 1 0 0 0 0 |
| − | 44 47 1 0 0 0 0 | + | 44 47 1 0 0 0 0 |
| − | 43 48 1 0 0 0 0 | + | 43 48 1 0 0 0 0 |
| − | 45 49 1 0 0 0 0 | + | 45 49 1 0 0 0 0 |
| − | 40 50 1 0 0 0 0 | + | 40 50 1 0 0 0 0 |
| − | 33 18 1 0 0 0 0 | + | 33 18 1 0 0 0 0 |
| − | 51 52 1 0 0 0 0 | + | 51 52 1 0 0 0 0 |
| − | 26 51 1 0 0 0 0 | + | 26 51 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 51 52 | + | M SAL 1 2 51 52 |
| − | M SBL 1 1 57 | + | M SBL 1 1 57 |
| − | M SMT 1 ^ CH2OH | + | M SMT 1 ^ CH2OH |
| − | M SBV 1 57 0.6474 -0.4618 | + | M SBV 1 57 0.6474 -0.4618 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL3FAAGS0030 | + | ID FL3FAAGS0030 |
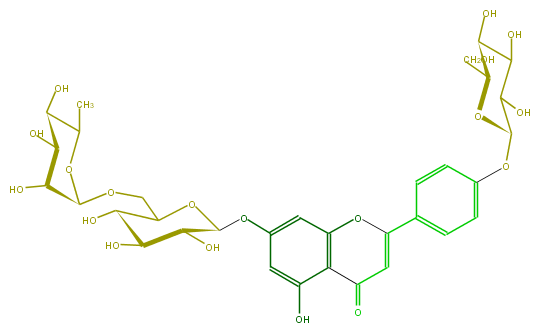
| − | FORMULA C33H40O19 | + | FORMULA C33H40O19 |
| − | EXACTMASS 740.216379098 | + | EXACTMASS 740.216379098 |
| − | AVERAGEMASS 740.6593 | + | AVERAGEMASS 740.6593 |
| − | SMILES Oc(c4)c(C(=O)3)c(cc4OC(C6O)OC(C(O)C6O)COC(C5O)OC(C)C(C(O)5)O)OC(=C3)c(c1)ccc(OC(O2)C(O)C(C(O)C(CO)2)O)c1 | + | SMILES Oc(c4)c(C(=O)3)c(cc4OC(C6O)OC(C(O)C6O)COC(C5O)OC(C)C(C(O)5)O)OC(=C3)c(c1)ccc(OC(O2)C(O)C(C(O)C(CO)2)O)c1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
52 57 0 0 0 0 0 0 0 0999 V2000
0.0049 -1.6485 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0049 -2.4581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7060 -2.8630 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4071 -2.4581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4071 -1.6485 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7060 -1.2437 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1082 -2.8630 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8094 -2.4581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8094 -1.6485 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1082 -1.2437 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1082 -3.4942 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5103 -1.2439 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2249 -1.6565 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.9393 -1.2439 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.9393 -0.4188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2249 -0.0062 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5103 -0.4188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6328 -1.2453 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7060 -3.6723 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.6539 -0.0063 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.9859 1.2063 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.7777 0.7491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.5265 1.6282 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.7777 2.5126 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.9859 2.9698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.2372 2.0908 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.1742 3.6723 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.7777 3.1695 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.9921 1.2864 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6935 -1.0739 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0437 -1.9317 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1079 -1.5678 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2048 -1.5581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8609 -0.9018 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6797 -1.3338 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0574 -0.7724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2420 -1.2928 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7027 -1.8919 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4540 -1.9452 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.3509 -0.4940 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.5591 -0.9512 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.8103 -0.0722 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.5591 0.8124 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.3509 1.2696 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.0996 0.3904 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8137 -0.6321 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.0879 1.8683 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.5591 1.4776 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.6874 0.7926 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.9921 -0.5573 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.5898 2.5526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6290 1.7367 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
3 19 1 0 0 0 0
15 20 1 0 0 0 0
22 21 1 1 0 0 0
22 23 1 0 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 26 1 1 0 0 0
26 21 1 1 0 0 0
25 27 1 0 0 0 0
24 28 1 0 0 0 0
23 29 1 0 0 0 0
22 20 1 0 0 0 0
30 31 1 1 0 0 0
31 32 1 1 0 0 0
33 32 1 1 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 30 1 0 0 0 0
35 36 1 0 0 0 0
30 37 1 0 0 0 0
31 38 1 0 0 0 0
32 39 1 0 0 0 0
41 40 1 1 0 0 0
41 42 1 0 0 0 0
42 43 1 0 0 0 0
43 44 1 0 0 0 0
44 45 1 1 0 0 0
45 40 1 1 0 0 0
41 46 1 0 0 0 0
46 36 1 0 0 0 0
44 47 1 0 0 0 0
43 48 1 0 0 0 0
45 49 1 0 0 0 0
40 50 1 0 0 0 0
33 18 1 0 0 0 0
51 52 1 0 0 0 0
26 51 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 51 52
M SBL 1 1 57
M SMT 1 ^ CH2OH
M SBV 1 57 0.6474 -0.4618
S SKP 5
ID FL3FAAGS0030
FORMULA C33H40O19
EXACTMASS 740.216379098
AVERAGEMASS 740.6593
SMILES Oc(c4)c(C(=O)3)c(cc4OC(C6O)OC(C(O)C6O)COC(C5O)OC(C)C(C(O)5)O)OC(=C3)c(c1)ccc(OC(O2)C(O)C(C(O)C(CO)2)O)c1
M END