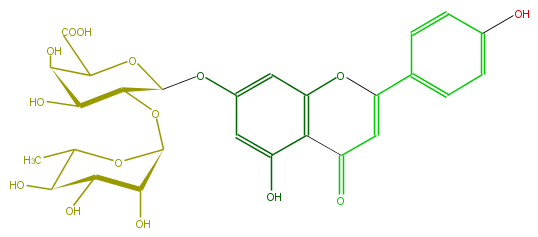
Mol:FL3FAAGS0024
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 42 46 0 0 0 0 0 0 0 0999 V2000 | + | 42 46 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.6714 0.1259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6714 0.1259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6714 -0.6836 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6714 -0.6836 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0297 -1.0883 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0297 -1.0883 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7307 -0.6836 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7307 -0.6836 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7307 0.1259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7307 0.1259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0297 0.5307 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0297 0.5307 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4317 -1.0883 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4317 -1.0883 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1327 -0.6836 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1327 -0.6836 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1327 0.1259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1327 0.1259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4317 0.5307 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4317 0.5307 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4317 -1.9642 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4317 -1.9642 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8335 0.5304 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8335 0.5304 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5480 0.1180 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5480 0.1180 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2625 0.5304 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2625 0.5304 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2625 1.3555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2625 1.3555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5480 1.7680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5480 1.7680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8335 1.3555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8335 1.3555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3721 0.5304 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3721 0.5304 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0297 -1.8975 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0297 -1.8975 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9767 1.7679 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9767 1.7679 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3834 0.6794 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3834 0.6794 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7985 -0.0926 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7985 -0.0926 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9564 0.2349 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9564 0.2349 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1439 0.2437 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1439 0.2437 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7343 0.8342 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7343 0.8342 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5946 0.5254 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5946 0.5254 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3821 1.0290 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3821 1.0290 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5850 0.0060 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5850 0.0060 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2798 -0.2277 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2798 -0.2277 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9019 -0.9626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9019 -0.9626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3616 -1.7587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3616 -1.7587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4776 -1.5061 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4776 -1.5061 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5882 -1.7587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5882 -1.7587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1284 -0.9626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1284 -0.9626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0125 -1.2151 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0125 -1.2151 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9950 -2.1707 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9950 -2.1707 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9767 -1.6582 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9767 -1.6582 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6317 -1.1440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6317 -1.1440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5882 -2.4297 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5882 -2.4297 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1827 1.4018 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1827 1.4018 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6620 2.4297 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6620 2.4297 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5323 2.3308 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5323 2.3308 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 30 1 0 0 0 0 | + | 35 30 1 0 0 0 0 |
| − | 34 29 1 0 0 0 0 | + | 34 29 1 0 0 0 0 |
| − | 32 36 1 0 0 0 0 | + | 32 36 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 30 38 1 0 0 0 0 | + | 30 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 24 18 1 0 0 0 0 | + | 24 18 1 0 0 0 0 |
| − | 40 41 2 0 0 0 0 | + | 40 41 2 0 0 0 0 |
| − | 40 42 1 0 0 0 0 | + | 40 42 1 0 0 0 0 |
| − | 26 40 1 0 0 0 0 | + | 26 40 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 3 40 41 42 | + | M SAL 1 3 40 41 42 |
| − | M SBL 1 1 46 | + | M SBL 1 1 46 |
| − | M SMT 1 ^ COOH | + | M SMT 1 ^ COOH |
| − | M SBV 1 46 0.5882 -0.8764 | + | M SBV 1 46 0.5882 -0.8764 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL3FAAGS0024 | + | ID FL3FAAGS0024 |
| − | FORMULA C27H28O15 | + | FORMULA C27H28O15 |
| − | EXACTMASS 592.1428202259999 | + | EXACTMASS 592.1428202259999 |
| − | AVERAGEMASS 592.5022200000001 | + | AVERAGEMASS 592.5022200000001 |
| − | SMILES c(c5O)c(cc(c45)OC(=CC4=O)c(c3)ccc(c3)O)OC(O1)C(OC(C2O)OC(C)C(C2O)O)C(C(C1C(O)=O)O)O | + | SMILES c(c5O)c(cc(c45)OC(=CC4=O)c(c3)ccc(c3)O)OC(O1)C(OC(C2O)OC(C)C(C2O)O)C(C(C1C(O)=O)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
42 46 0 0 0 0 0 0 0 0999 V2000
-0.6714 0.1259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6714 -0.6836 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0297 -1.0883 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7307 -0.6836 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7307 0.1259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0297 0.5307 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4317 -1.0883 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1327 -0.6836 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1327 0.1259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4317 0.5307 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4317 -1.9642 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8335 0.5304 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5480 0.1180 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2625 0.5304 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2625 1.3555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5480 1.7680 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8335 1.3555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3721 0.5304 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0297 -1.8975 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.9767 1.7679 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.3834 0.6794 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7985 -0.0926 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9564 0.2349 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1439 0.2437 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7343 0.8342 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5946 0.5254 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3821 1.0290 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.5850 0.0060 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2798 -0.2277 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9019 -0.9626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3616 -1.7587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4776 -1.5061 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5882 -1.7587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1284 -0.9626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0125 -1.2151 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9950 -2.1707 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.9767 -1.6582 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.6317 -1.1440 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5882 -2.4297 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1827 1.4018 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.6620 2.4297 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5323 2.3308 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
3 19 1 0 0 0 0
15 20 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
30 31 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 30 1 0 0 0 0
34 29 1 0 0 0 0
32 36 1 0 0 0 0
31 37 1 0 0 0 0
30 38 1 0 0 0 0
33 39 1 0 0 0 0
24 18 1 0 0 0 0
40 41 2 0 0 0 0
40 42 1 0 0 0 0
26 40 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 3 40 41 42
M SBL 1 1 46
M SMT 1 ^ COOH
M SBV 1 46 0.5882 -0.8764
S SKP 5
ID FL3FAAGS0024
FORMULA C27H28O15
EXACTMASS 592.1428202259999
AVERAGEMASS 592.5022200000001
SMILES c(c5O)c(cc(c45)OC(=CC4=O)c(c3)ccc(c3)O)OC(O1)C(OC(C2O)OC(C)C(C2O)O)C(C(C1C(O)=O)O)O
M END