Mol:FL3FAAGS0002
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 40 44 0 0 0 0 0 0 0 0999 V2000 | + | 40 44 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.3935 0.3858 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3935 0.3858 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3935 -0.3926 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3935 -0.3926 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2807 -0.7819 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2807 -0.7819 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9548 -0.3926 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9548 -0.3926 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9548 0.3858 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9548 0.3858 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2807 0.7752 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2807 0.7752 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6291 -0.7819 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6291 -0.7819 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3032 -0.3926 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3032 -0.3926 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3032 0.3858 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3032 0.3858 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6291 0.7752 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6291 0.7752 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9769 0.7749 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9769 0.7749 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6914 0.3624 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6914 0.3624 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4058 0.7749 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4058 0.7749 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4058 1.5999 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4058 1.5999 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6914 2.0124 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6914 2.0124 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9769 1.5999 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9769 1.5999 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2807 -1.5599 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2807 -1.5599 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0673 0.7749 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0673 0.7749 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1197 2.0120 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1197 2.0120 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6291 -1.5593 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6291 -1.5593 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4207 0.6961 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4207 0.6961 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7399 0.0152 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7399 0.0152 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9031 0.4920 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9031 0.4920 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9405 0.4920 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9405 0.4920 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6212 1.1730 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6212 1.1730 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4579 0.6961 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4579 0.6961 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0072 -0.0327 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0072 -0.0327 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.1197 0.4022 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.1197 0.4022 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5269 0.0345 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5269 0.0345 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0041 -0.7404 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0041 -0.7404 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0223 -0.3005 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0223 -0.3005 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0741 -0.7749 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0741 -0.7749 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4966 -1.4349 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4966 -1.4349 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5066 -1.3936 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5066 -1.3936 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4966 -1.9921 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4966 -1.9921 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5066 -2.0124 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5066 -2.0124 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1882 1.1377 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1882 1.1377 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1882 1.9336 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1882 1.9336 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4966 -0.8148 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4966 -0.8148 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8438 -0.8148 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8438 -0.8148 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 9 11 1 0 0 0 0 | + | 9 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 3 17 1 0 0 0 0 | + | 3 17 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 14 19 1 0 0 0 0 | + | 14 19 1 0 0 0 0 |
| − | 7 20 2 0 0 0 0 | + | 7 20 2 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 24 18 1 0 0 0 0 | + | 24 18 1 0 0 0 0 |
| − | 23 27 1 0 0 0 0 | + | 23 27 1 0 0 0 0 |
| − | 21 28 1 0 0 0 0 | + | 21 28 1 0 0 0 0 |
| − | 22 29 1 0 0 0 0 | + | 22 29 1 0 0 0 0 |
| − | 27 30 1 0 0 0 0 | + | 27 30 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 30 34 1 1 0 0 0 | + | 30 34 1 1 0 0 0 |
| − | 33 35 1 0 0 0 0 | + | 33 35 1 0 0 0 0 |
| − | 34 36 1 0 0 0 0 | + | 34 36 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 26 37 1 0 0 0 0 | + | 26 37 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 37 38 | + | M SAL 1 2 37 38 |
| − | M SBL 1 1 42 | + | M SBL 1 1 42 |
| − | M SMT 1 ^ CH2OH | + | M SMT 1 ^ CH2OH |
| − | M SBV 1 42 0.7303 -0.4416 | + | M SBV 1 42 0.7303 -0.4416 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 39 40 | + | M SAL 2 2 39 40 |
| − | M SBL 2 1 44 | + | M SBL 2 1 44 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SBV 2 44 0.0000 -0.6201 | + | M SBV 2 44 0.0000 -0.6201 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL3FAAGS0002 | + | ID FL3FAAGS0002 |
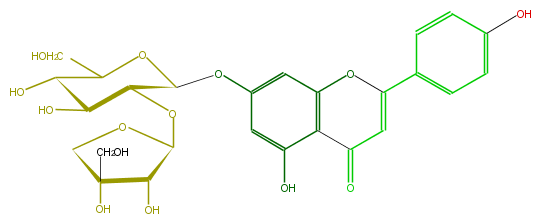
| − | FORMULA C26H28O14 | + | FORMULA C26H28O14 |
| − | EXACTMASS 564.147905604 | + | EXACTMASS 564.147905604 |
| − | AVERAGEMASS 564.49212 | + | AVERAGEMASS 564.49212 |
| − | SMILES c(c13)c(OC(O4)C(OC(O5)C(O)C(O)(C5)CO)C(O)C(C(CO)4)O)cc(c(C(C=C(O3)c(c2)ccc(c2)O)=O)1)O | + | SMILES c(c13)c(OC(O4)C(OC(O5)C(O)C(O)(C5)CO)C(O)C(C(CO)4)O)cc(c(C(C=C(O3)c(c2)ccc(c2)O)=O)1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
40 44 0 0 0 0 0 0 0 0999 V2000
-0.3935 0.3858 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3935 -0.3926 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2807 -0.7819 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9548 -0.3926 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9548 0.3858 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2807 0.7752 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6291 -0.7819 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3032 -0.3926 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3032 0.3858 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6291 0.7752 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9769 0.7749 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6914 0.3624 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4058 0.7749 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4058 1.5999 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6914 2.0124 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9769 1.5999 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2807 -1.5599 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0673 0.7749 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.1197 2.0120 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6291 -1.5593 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.4207 0.6961 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7399 0.0152 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9031 0.4920 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9405 0.4920 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6212 1.1730 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4579 0.6961 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0072 -0.0327 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.1197 0.4022 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.5269 0.0345 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0041 -0.7404 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0223 -0.3005 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0741 -0.7749 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4966 -1.4349 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5066 -1.3936 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4966 -1.9921 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5066 -2.0124 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1882 1.1377 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1882 1.9336 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4966 -0.8148 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.8438 -0.8148 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
3 17 1 0 0 0 0
1 18 1 0 0 0 0
14 19 1 0 0 0 0
7 20 2 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
24 18 1 0 0 0 0
23 27 1 0 0 0 0
21 28 1 0 0 0 0
22 29 1 0 0 0 0
27 30 1 0 0 0 0
30 31 1 0 0 0 0
31 32 1 0 0 0 0
32 33 1 1 0 0 0
33 34 1 1 0 0 0
30 34 1 1 0 0 0
33 35 1 0 0 0 0
34 36 1 0 0 0 0
37 38 1 0 0 0 0
26 37 1 0 0 0 0
39 40 1 0 0 0 0
33 39 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 37 38
M SBL 1 1 42
M SMT 1 ^ CH2OH
M SBV 1 42 0.7303 -0.4416
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 39 40
M SBL 2 1 44
M SMT 2 CH2OH
M SBV 2 44 0.0000 -0.6201
S SKP 5
ID FL3FAAGS0002
FORMULA C26H28O14
EXACTMASS 564.147905604
AVERAGEMASS 564.49212
SMILES c(c13)c(OC(O4)C(OC(O5)C(O)C(O)(C5)CO)C(O)C(C(CO)4)O)cc(c(C(C=C(O3)c(c2)ccc(c2)O)=O)1)O
M END