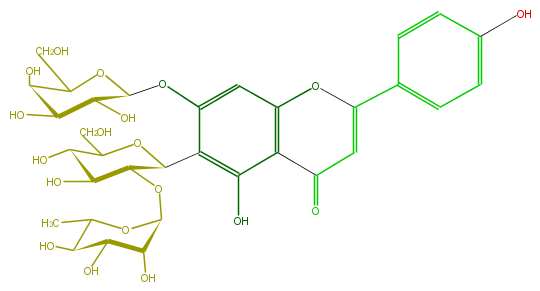
Mol:FL3FAADS0029
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 52 57 0 0 0 0 0 0 0 0999 V2000 | + | 52 57 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.4731 0.7383 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4731 0.7383 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4731 -0.1860 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4731 -0.1860 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6727 -0.6482 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6727 -0.6482 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1277 -0.1860 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1277 -0.1860 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1277 0.7383 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1277 0.7383 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6727 1.2002 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6727 1.2002 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9281 -0.6482 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9281 -0.6482 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7285 -0.1860 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7285 -0.1860 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7285 0.7383 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7285 0.7383 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9281 1.2002 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9281 1.2002 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9281 -1.3687 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9281 -1.3687 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2166 1.2443 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2166 1.2443 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6727 -1.5720 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6727 -1.5720 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6177 1.2299 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6177 1.2299 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4611 0.7429 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4611 0.7429 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3045 1.2299 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3045 1.2299 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3045 2.2038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3045 2.2038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4611 2.6907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4611 2.6907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6177 2.2038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6177 2.2038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1475 2.6904 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1475 2.6904 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1421 -0.1094 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1421 -0.1094 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6440 -0.7671 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6440 -0.7671 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9266 -0.4881 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9266 -0.4881 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1789 -0.4962 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1789 -0.4962 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7374 0.0226 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7374 0.0226 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4702 -0.2406 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4702 -0.2406 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6759 -0.3166 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6759 -0.3166 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3779 -0.7684 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3779 -0.7684 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3021 -0.9105 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3021 -0.9105 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7007 -1.5568 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7007 -1.5568 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0752 -2.2054 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0752 -2.2054 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3549 -1.9997 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3549 -1.9997 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6304 -2.2054 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6304 -2.2054 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2557 -1.5568 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2557 -1.5568 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9761 -1.7625 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9761 -1.7625 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4177 -1.6194 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4177 -1.6194 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5296 -2.0836 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5296 -2.0836 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7597 -2.6140 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7597 -2.6140 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5849 -2.7560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5849 -2.7560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9785 1.2086 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9785 1.2086 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3849 0.5530 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3849 0.5530 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6449 0.9179 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6449 0.9179 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9064 0.9905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9064 0.9905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4907 1.4807 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4907 1.4807 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1298 1.0684 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1298 1.0684 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9551 1.5206 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9551 1.5206 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.1475 0.6038 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.1475 0.6038 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0050 0.5552 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0050 0.5552 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9626 0.2519 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9626 0.2519 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2699 1.3985 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2699 1.3985 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8621 1.9246 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8621 1.9246 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6394 2.7560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6394 2.7560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 1 12 1 0 0 0 0 | + | 1 12 1 0 0 0 0 |
| − | 3 13 1 0 0 0 0 | + | 3 13 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 18 1 0 0 0 0 | + | 17 18 1 0 0 0 0 |
| − | 18 19 2 0 0 0 0 | + | 18 19 2 0 0 0 0 |
| − | 19 14 1 0 0 0 0 | + | 19 14 1 0 0 0 0 |
| − | 14 9 1 0 0 0 0 | + | 14 9 1 0 0 0 0 |
| − | 17 20 1 0 0 0 0 | + | 17 20 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 30 1 0 0 0 0 | + | 35 30 1 0 0 0 0 |
| − | 30 36 1 0 0 0 0 | + | 30 36 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 34 29 1 0 0 0 0 | + | 34 29 1 0 0 0 0 |
| − | 24 2 1 0 0 0 0 | + | 24 2 1 0 0 0 0 |
| − | 40 41 1 1 0 0 0 | + | 40 41 1 1 0 0 0 |
| − | 41 42 1 1 0 0 0 | + | 41 42 1 1 0 0 0 |
| − | 43 42 1 1 0 0 0 | + | 43 42 1 1 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 45 40 1 0 0 0 0 | + | 45 40 1 0 0 0 0 |
| − | 40 46 1 0 0 0 0 | + | 40 46 1 0 0 0 0 |
| − | 41 47 1 0 0 0 0 | + | 41 47 1 0 0 0 0 |
| − | 42 48 1 0 0 0 0 | + | 42 48 1 0 0 0 0 |
| − | 43 12 1 0 0 0 0 | + | 43 12 1 0 0 0 0 |
| − | 49 50 1 0 0 0 0 | + | 49 50 1 0 0 0 0 |
| − | 26 49 1 0 0 0 0 | + | 26 49 1 0 0 0 0 |
| − | 51 52 1 0 0 0 0 | + | 51 52 1 0 0 0 0 |
| − | 45 51 1 0 0 0 0 | + | 45 51 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 49 50 | + | M SAL 1 2 49 50 |
| − | M SBL 1 1 55 | + | M SBL 1 1 55 |
| − | M SMT 1 ^ CH2OH | + | M SMT 1 ^ CH2OH |
| − | M SBV 1 55 0.4924 -0.4925 | + | M SBV 1 55 0.4924 -0.4925 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 51 52 | + | M SAL 2 2 51 52 |
| − | M SBL 2 1 57 | + | M SBL 2 1 57 |
| − | M SMT 2 ^ CH2OH | + | M SMT 2 ^ CH2OH |
| − | M SBV 2 57 0.7324 -0.8561 | + | M SBV 2 57 0.7324 -0.8561 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL3FAADS0029 | + | ID FL3FAADS0029 |
| − | FORMULA C33H40O19 | + | FORMULA C33H40O19 |
| − | EXACTMASS 740.216379098 | + | EXACTMASS 740.216379098 |
| − | AVERAGEMASS 740.6593 | + | AVERAGEMASS 740.6593 |
| − | SMILES c(c(C(O5)C(OC(C6O)OC(C)C(C6O)O)C(O)C(C5CO)O)3)(OC(O4)C(C(O)C(O)C4CO)O)cc(c2c(O)3)OC(=CC2=O)c(c1)ccc(O)c1 | + | SMILES c(c(C(O5)C(OC(C6O)OC(C)C(C6O)O)C(O)C(C5CO)O)3)(OC(O4)C(C(O)C(O)C4CO)O)cc(c2c(O)3)OC(=CC2=O)c(c1)ccc(O)c1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
52 57 0 0 0 0 0 0 0 0999 V2000
-1.4731 0.7383 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4731 -0.1860 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6727 -0.6482 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1277 -0.1860 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1277 0.7383 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6727 1.2002 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9281 -0.6482 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7285 -0.1860 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7285 0.7383 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9281 1.2002 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9281 -1.3687 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2166 1.2443 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6727 -1.5720 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6177 1.2299 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4611 0.7429 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3045 1.2299 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3045 2.2038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4611 2.6907 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6177 2.2038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.1475 2.6904 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1421 -0.1094 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6440 -0.7671 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9266 -0.4881 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1789 -0.4962 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7374 0.0226 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4702 -0.2406 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.6759 -0.3166 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.3779 -0.7684 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3021 -0.9105 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7007 -1.5568 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0752 -2.2054 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3549 -1.9997 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6304 -2.2054 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2557 -1.5568 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9761 -1.7625 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.4177 -1.6194 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.5296 -2.0836 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7597 -2.6140 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5849 -2.7560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.9785 1.2086 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3849 0.5530 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6449 0.9179 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9064 0.9905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4907 1.4807 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1298 1.0684 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.9551 1.5206 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.1475 0.6038 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0050 0.5552 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9626 0.2519 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2699 1.3985 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.8621 1.9246 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.6394 2.7560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
1 12 1 0 0 0 0
3 13 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 18 1 0 0 0 0
18 19 2 0 0 0 0
19 14 1 0 0 0 0
14 9 1 0 0 0 0
17 20 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
30 31 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 30 1 0 0 0 0
30 36 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
34 29 1 0 0 0 0
24 2 1 0 0 0 0
40 41 1 1 0 0 0
41 42 1 1 0 0 0
43 42 1 1 0 0 0
43 44 1 0 0 0 0
44 45 1 0 0 0 0
45 40 1 0 0 0 0
40 46 1 0 0 0 0
41 47 1 0 0 0 0
42 48 1 0 0 0 0
43 12 1 0 0 0 0
49 50 1 0 0 0 0
26 49 1 0 0 0 0
51 52 1 0 0 0 0
45 51 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 49 50
M SBL 1 1 55
M SMT 1 ^ CH2OH
M SBV 1 55 0.4924 -0.4925
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 51 52
M SBL 2 1 57
M SMT 2 ^ CH2OH
M SBV 2 57 0.7324 -0.8561
S SKP 5
ID FL3FAADS0029
FORMULA C33H40O19
EXACTMASS 740.216379098
AVERAGEMASS 740.6593
SMILES c(c(C(O5)C(OC(C6O)OC(C)C(C6O)O)C(O)C(C5CO)O)3)(OC(O4)C(C(O)C(O)C4CO)O)cc(c2c(O)3)OC(=CC2=O)c(c1)ccc(O)c1
M END