Mol:FL3FAADS0027
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 42 46 0 0 0 0 0 0 0 0999 V2000 | + | 42 46 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.6659 -1.2287 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6659 -1.2287 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6659 -1.8710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6659 -1.8710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1096 -2.1922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1096 -2.1922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5533 -1.8710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5533 -1.8710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5533 -1.2287 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5533 -1.2287 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1096 -0.9075 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1096 -0.9075 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0030 -2.1922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0030 -2.1922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5593 -1.8710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5593 -1.8710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5593 -1.2287 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5593 -1.2287 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0030 -0.9075 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0030 -0.9075 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0030 -2.6931 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0030 -2.6931 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2220 -0.9076 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2220 -0.9076 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1096 -2.8343 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1096 -2.8343 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1772 -0.8870 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1772 -0.8870 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7635 -1.2254 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7635 -1.2254 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3497 -0.8870 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3497 -0.8870 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3497 -0.2101 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3497 -0.2101 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7635 0.1284 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7635 0.1284 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1772 -0.2101 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1772 -0.2101 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9355 0.1282 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9355 0.1282 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1783 2.5607 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1783 2.5607 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5727 2.1019 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5727 2.1019 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8296 1.4413 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8296 1.4413 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8365 0.8038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8365 0.8038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2998 1.2671 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2998 1.2671 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9948 1.8450 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9948 1.8450 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5379 2.8171 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5379 2.8171 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1942 1.0628 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1942 1.0628 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6190 -2.1598 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6190 -2.1598 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2478 -2.6497 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2478 -2.6497 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7133 -2.4419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7133 -2.4419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1976 -2.4363 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1976 -2.4363 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5724 -2.0615 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5724 -2.0615 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0400 -2.3083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0400 -2.3083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1329 -2.4564 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1329 -2.4564 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8265 -2.6779 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8265 -2.6779 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4071 -2.9560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4071 -2.9560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0724 2.9560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0724 2.9560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4184 2.0896 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4184 2.0896 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1329 1.6771 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1329 1.6771 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2966 -1.3102 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2966 -1.3102 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5821 -1.7227 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5821 -1.7227 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 1 12 1 0 0 0 0 | + | 1 12 1 0 0 0 0 |
| − | 3 13 1 0 0 0 0 | + | 3 13 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 18 1 0 0 0 0 | + | 17 18 1 0 0 0 0 |
| − | 18 19 2 0 0 0 0 | + | 18 19 2 0 0 0 0 |
| − | 19 14 1 0 0 0 0 | + | 19 14 1 0 0 0 0 |
| − | 14 9 1 0 0 0 0 | + | 14 9 1 0 0 0 0 |
| − | 17 20 1 0 0 0 0 | + | 17 20 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 22 27 1 0 0 0 0 | + | 22 27 1 0 0 0 0 |
| − | 23 28 1 0 0 0 0 | + | 23 28 1 0 0 0 0 |
| − | 24 20 1 0 0 0 0 | + | 24 20 1 0 0 0 0 |
| − | 29 30 1 1 0 0 0 | + | 29 30 1 1 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 32 31 1 1 0 0 0 | + | 32 31 1 1 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 29 1 0 0 0 0 | + | 34 29 1 0 0 0 0 |
| − | 29 35 1 0 0 0 0 | + | 29 35 1 0 0 0 0 |
| − | 30 36 1 0 0 0 0 | + | 30 36 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 2 32 1 0 0 0 0 | + | 2 32 1 0 0 0 0 |
| − | 21 38 1 0 0 0 0 | + | 21 38 1 0 0 0 0 |
| − | 26 39 1 0 0 0 0 | + | 26 39 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 34 41 1 0 0 0 0 | + | 34 41 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 39 40 | + | M SAL 1 2 39 40 |
| − | M SBL 1 1 43 | + | M SBL 1 1 43 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 43 -6.8894 5.3272 | + | M SBV 1 43 -6.8894 5.3272 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 41 42 | + | M SAL 2 2 41 42 |
| − | M SBL 2 1 45 | + | M SBL 2 1 45 |
| − | M SMT 2 ^CH2OH | + | M SMT 2 ^CH2OH |
| − | M SBV 2 45 -7.5697 6.0806 | + | M SBV 2 45 -7.5697 6.0806 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL3FAADS0027 | + | ID FL3FAADS0027 |
| − | KNApSAcK_ID C00006393 | + | KNApSAcK_ID C00006393 |
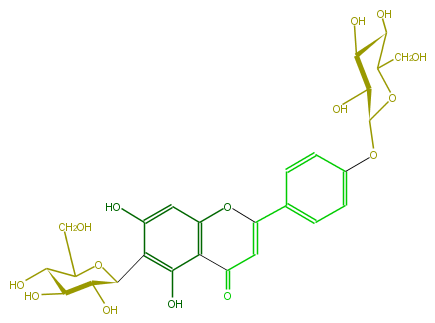
| − | NAME Isosaponarin | + | NAME Isosaponarin |
| − | CAS_RN 19416-87-6 | + | CAS_RN 19416-87-6 |
| − | FORMULA C27H30O15 | + | FORMULA C27H30O15 |
| − | EXACTMASS 594.15847029 | + | EXACTMASS 594.15847029 |
| − | AVERAGEMASS 594.5181 | + | AVERAGEMASS 594.5181 |
| − | SMILES C(c(c5O)c(O)cc(c52)OC(c(c3)ccc(OC(O4)C(O)C(O)C(O)C4CO)c3)=CC2=O)(O1)C(C(O)C(O)C(CO)1)O | + | SMILES C(c(c5O)c(O)cc(c52)OC(c(c3)ccc(OC(O4)C(O)C(O)C(O)C4CO)c3)=CC2=O)(O1)C(C(O)C(O)C(CO)1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
42 46 0 0 0 0 0 0 0 0999 V2000
-1.6659 -1.2287 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6659 -1.8710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1096 -2.1922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5533 -1.8710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5533 -1.2287 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1096 -0.9075 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0030 -2.1922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5593 -1.8710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5593 -1.2287 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0030 -0.9075 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0030 -2.6931 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2220 -0.9076 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.1096 -2.8343 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1772 -0.8870 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7635 -1.2254 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3497 -0.8870 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3497 -0.2101 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7635 0.1284 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1772 -0.2101 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9355 0.1282 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1783 2.5607 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5727 2.1019 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8296 1.4413 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8365 0.8038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2998 1.2671 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9948 1.8450 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5379 2.8171 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1942 1.0628 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6190 -2.1598 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2478 -2.6497 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7133 -2.4419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1976 -2.4363 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5724 -2.0615 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0400 -2.3083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1329 -2.4564 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.8265 -2.6779 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4071 -2.9560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0724 2.9560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4184 2.0896 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1329 1.6771 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2966 -1.3102 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5821 -1.7227 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
1 12 1 0 0 0 0
3 13 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 18 1 0 0 0 0
18 19 2 0 0 0 0
19 14 1 0 0 0 0
14 9 1 0 0 0 0
17 20 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
22 27 1 0 0 0 0
23 28 1 0 0 0 0
24 20 1 0 0 0 0
29 30 1 1 0 0 0
30 31 1 1 0 0 0
32 31 1 1 0 0 0
32 33 1 0 0 0 0
33 34 1 0 0 0 0
34 29 1 0 0 0 0
29 35 1 0 0 0 0
30 36 1 0 0 0 0
31 37 1 0 0 0 0
2 32 1 0 0 0 0
21 38 1 0 0 0 0
26 39 1 0 0 0 0
39 40 1 0 0 0 0
34 41 1 0 0 0 0
41 42 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 39 40
M SBL 1 1 43
M SMT 1 CH2OH
M SBV 1 43 -6.8894 5.3272
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 41 42
M SBL 2 1 45
M SMT 2 ^CH2OH
M SBV 2 45 -7.5697 6.0806
S SKP 8
ID FL3FAADS0027
KNApSAcK_ID C00006393
NAME Isosaponarin
CAS_RN 19416-87-6
FORMULA C27H30O15
EXACTMASS 594.15847029
AVERAGEMASS 594.5181
SMILES C(c(c5O)c(O)cc(c52)OC(c(c3)ccc(OC(O4)C(O)C(O)C(O)C4CO)c3)=CC2=O)(O1)C(C(O)C(O)C(CO)1)O
M END