Mol:FL3FAADS0015
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 52 57 0 0 0 0 0 0 0 0999 V2000 | + | 52 57 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.1187 1.0153 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1187 1.0153 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1187 0.0659 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1187 0.0659 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2964 -0.4089 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2964 -0.4089 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5258 0.0659 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5258 0.0659 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5258 1.0153 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5258 1.0153 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2964 1.4899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2964 1.4899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3480 -0.4089 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3480 -0.4089 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1702 0.0659 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1702 0.0659 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1702 1.0153 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1702 1.0153 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3480 1.4899 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3480 1.4899 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3480 -1.1490 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3480 -1.1490 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8697 1.4363 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8697 1.4363 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2964 -1.3579 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2964 -1.3579 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0834 1.5203 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0834 1.5203 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9498 1.0200 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9498 1.0200 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8164 1.5203 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8164 1.5203 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8164 2.5208 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8164 2.5208 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9498 3.0209 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9498 3.0209 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0834 2.5208 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0834 2.5208 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.6822 3.0206 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.6822 3.0206 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9107 0.0258 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9107 0.0258 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3989 -0.6498 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3989 -0.6498 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6619 -0.3632 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6619 -0.3632 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8940 -0.3717 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8940 -0.3717 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4676 0.1613 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4676 0.1613 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2205 -0.1090 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2205 -0.1090 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4814 -0.2030 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4814 -0.2030 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1967 -0.6887 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1967 -0.6887 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9703 -0.9497 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9703 -0.9497 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3590 -1.7054 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3590 -1.7054 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7437 -2.3717 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7437 -2.3717 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0040 -2.1603 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0040 -2.1603 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2596 -2.3717 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2596 -2.3717 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8748 -1.7054 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8748 -1.7054 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6147 -1.9167 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6147 -1.9167 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1341 -1.8363 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1341 -1.8363 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1768 -2.3112 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1768 -2.3112 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5211 -2.7534 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5211 -2.7534 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2596 -3.0209 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2596 -3.0209 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6637 1.5605 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6637 1.5605 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1521 0.8850 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1521 0.8850 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4151 1.1715 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4151 1.1715 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6470 1.1631 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6470 1.1631 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2208 1.6961 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2208 1.6961 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9735 1.4258 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9735 1.4258 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.2360 1.2301 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.2360 1.2301 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9497 0.8461 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9497 0.8461 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6821 0.6676 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6821 0.6676 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7925 0.4165 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7925 0.4165 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9703 0.7322 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9703 0.7322 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5044 1.9720 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5044 1.9720 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.6822 2.2877 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.6822 2.2877 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 1 12 1 0 0 0 0 | + | 1 12 1 0 0 0 0 |
| − | 3 13 1 0 0 0 0 | + | 3 13 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 18 1 0 0 0 0 | + | 17 18 1 0 0 0 0 |
| − | 18 19 2 0 0 0 0 | + | 18 19 2 0 0 0 0 |
| − | 19 14 1 0 0 0 0 | + | 19 14 1 0 0 0 0 |
| − | 14 9 1 0 0 0 0 | + | 14 9 1 0 0 0 0 |
| − | 17 20 1 0 0 0 0 | + | 17 20 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 30 1 0 0 0 0 | + | 35 30 1 0 0 0 0 |
| − | 30 36 1 0 0 0 0 | + | 30 36 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 34 29 1 0 0 0 0 | + | 34 29 1 0 0 0 0 |
| − | 24 2 1 0 0 0 0 | + | 24 2 1 0 0 0 0 |
| − | 40 41 1 1 0 0 0 | + | 40 41 1 1 0 0 0 |
| − | 41 42 1 1 0 0 0 | + | 41 42 1 1 0 0 0 |
| − | 43 42 1 1 0 0 0 | + | 43 42 1 1 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 45 40 1 0 0 0 0 | + | 45 40 1 0 0 0 0 |
| − | 40 46 1 0 0 0 0 | + | 40 46 1 0 0 0 0 |
| − | 41 47 1 0 0 0 0 | + | 41 47 1 0 0 0 0 |
| − | 42 48 1 0 0 0 0 | + | 42 48 1 0 0 0 0 |
| − | 43 12 1 0 0 0 0 | + | 43 12 1 0 0 0 0 |
| − | 49 50 1 0 0 0 0 | + | 49 50 1 0 0 0 0 |
| − | 26 49 1 0 0 0 0 | + | 26 49 1 0 0 0 0 |
| − | 51 52 1 0 0 0 0 | + | 51 52 1 0 0 0 0 |
| − | 45 51 1 0 0 0 0 | + | 45 51 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 49 50 | + | M SAL 1 2 49 50 |
| − | M SBL 1 1 55 | + | M SBL 1 1 55 |
| − | M SMT 1 ^CH2OH | + | M SMT 1 ^CH2OH |
| − | M SBV 1 55 0.5721 -0.5256 | + | M SBV 1 55 0.5721 -0.5256 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 51 52 | + | M SAL 2 2 51 52 |
| − | M SBL 2 1 57 | + | M SBL 2 1 57 |
| − | M SMT 2 ^CH2OH | + | M SMT 2 ^CH2OH |
| − | M SBV 2 57 0.5308 -0.5462 | + | M SBV 2 57 0.5308 -0.5462 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL3FAADS0015 | + | ID FL3FAADS0015 |
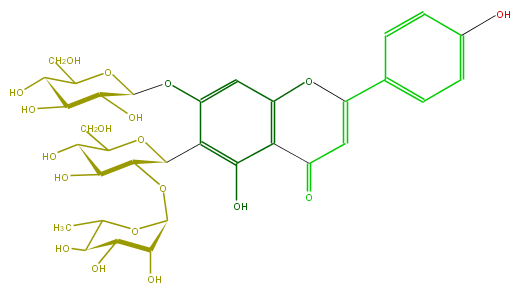
| − | FORMULA C33H40O19 | + | FORMULA C33H40O19 |
| − | EXACTMASS 740.216379098 | + | EXACTMASS 740.216379098 |
| − | AVERAGEMASS 740.6593 | + | AVERAGEMASS 740.6593 |
| − | SMILES c(c(C(O5)C(OC(C6O)OC(C)C(C6O)O)C(O)C(C5CO)O)3)(OC(O4)C(C(O)C(O)C4CO)O)cc(c2c(O)3)OC(=CC2=O)c(c1)ccc(O)c1 | + | SMILES c(c(C(O5)C(OC(C6O)OC(C)C(C6O)O)C(O)C(C5CO)O)3)(OC(O4)C(C(O)C(O)C4CO)O)cc(c2c(O)3)OC(=CC2=O)c(c1)ccc(O)c1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
52 57 0 0 0 0 0 0 0 0999 V2000
-1.1187 1.0153 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1187 0.0659 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2964 -0.4089 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5258 0.0659 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5258 1.0153 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2964 1.4899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3480 -0.4089 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1702 0.0659 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1702 1.0153 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3480 1.4899 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3480 -1.1490 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8697 1.4363 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2964 -1.3579 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0834 1.5203 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9498 1.0200 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.8164 1.5203 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.8164 2.5208 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9498 3.0209 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0834 2.5208 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.6822 3.0206 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9107 0.0258 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3989 -0.6498 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6619 -0.3632 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8940 -0.3717 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4676 0.1613 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2205 -0.1090 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4814 -0.2030 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1967 -0.6887 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9703 -0.9497 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3590 -1.7054 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7437 -2.3717 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0040 -2.1603 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2596 -2.3717 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8748 -1.7054 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6147 -1.9167 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1341 -1.8363 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1768 -2.3112 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5211 -2.7534 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2596 -3.0209 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.6637 1.5605 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1521 0.8850 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4151 1.1715 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6470 1.1631 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2208 1.6961 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9735 1.4258 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.2360 1.2301 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.9497 0.8461 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6821 0.6676 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7925 0.4165 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.9703 0.7322 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.5044 1.9720 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.6822 2.2877 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
1 12 1 0 0 0 0
3 13 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 18 1 0 0 0 0
18 19 2 0 0 0 0
19 14 1 0 0 0 0
14 9 1 0 0 0 0
17 20 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
30 31 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 30 1 0 0 0 0
30 36 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
34 29 1 0 0 0 0
24 2 1 0 0 0 0
40 41 1 1 0 0 0
41 42 1 1 0 0 0
43 42 1 1 0 0 0
43 44 1 0 0 0 0
44 45 1 0 0 0 0
45 40 1 0 0 0 0
40 46 1 0 0 0 0
41 47 1 0 0 0 0
42 48 1 0 0 0 0
43 12 1 0 0 0 0
49 50 1 0 0 0 0
26 49 1 0 0 0 0
51 52 1 0 0 0 0
45 51 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 49 50
M SBL 1 1 55
M SMT 1 ^CH2OH
M SBV 1 55 0.5721 -0.5256
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 51 52
M SBL 2 1 57
M SMT 2 ^CH2OH
M SBV 2 57 0.5308 -0.5462
S SKP 5
ID FL3FAADS0015
FORMULA C33H40O19
EXACTMASS 740.216379098
AVERAGEMASS 740.6593
SMILES c(c(C(O5)C(OC(C6O)OC(C)C(C6O)O)C(O)C(C5CO)O)3)(OC(O4)C(C(O)C(O)C4CO)O)cc(c2c(O)3)OC(=CC2=O)c(c1)ccc(O)c1
M END