Mol:FL2FEAGS0003
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 44 48 0 0 0 0 0 0 0 0999 V2000 | + | 44 48 0 0 0 0 0 0 0 0999 V2000 |
| − | 0.4814 0.6081 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4814 0.6081 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1941 1.0238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1941 1.0238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4846 -0.2169 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4846 -0.2169 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2014 -0.6262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2014 -0.6262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9140 -0.2105 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9140 -0.2105 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9104 0.6144 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9104 0.6144 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6303 -0.6199 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6303 -0.6199 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3429 -0.2042 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3429 -0.2042 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3393 0.6208 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3393 0.6208 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6230 1.0301 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6230 1.0301 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9931 1.0016 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9931 1.0016 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7091 0.5919 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7091 0.5919 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.4220 1.0071 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.4220 1.0071 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.4188 1.8321 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.4188 1.8321 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7028 2.2419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7028 2.2419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9899 1.8266 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9899 1.8266 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6314 -1.2631 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6314 -1.2631 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1440 0.9692 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1440 0.9692 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.0747 2.2108 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.0747 2.2108 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2014 -1.3343 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2014 -1.3343 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2323 -0.6308 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2323 -0.6308 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8158 1.1377 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8158 1.1377 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4032 0.4230 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4032 0.4230 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6048 0.6321 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6048 0.6321 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8113 0.4053 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8113 0.4053 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2239 1.1200 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2239 1.1200 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0223 0.9110 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0223 0.9110 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1793 1.4968 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1793 1.4968 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6591 1.7738 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6591 1.7738 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2990 0.6545 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2990 0.6545 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8518 0.6820 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8518 0.6820 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7763 -0.0078 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7763 -0.0078 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1963 -0.8139 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1963 -0.8139 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7856 -1.5252 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7856 -1.5252 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0202 -0.8139 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0202 -0.8139 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4321 -1.5274 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4321 -1.5274 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2560 -1.5274 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2560 -1.5274 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6685 -0.8129 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6685 -0.8129 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.4935 -0.8129 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.4935 -0.8129 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.9060 -1.5274 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.9060 -1.5274 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.4935 -2.2419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.4935 -2.2419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6685 -2.2419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6685 -2.2419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.7299 -1.5274 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.7299 -1.5274 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.7299 1.8326 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.7299 1.8326 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 1 3 1 0 0 0 0 | + | 1 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 2 1 0 0 0 0 | + | 6 2 1 0 0 0 0 |
| − | 5 7 1 0 0 0 0 | + | 5 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 6 1 0 0 0 0 | + | 10 6 1 0 0 0 0 |
| − | 9 11 1 0 0 0 0 | + | 9 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 7 17 2 0 0 0 0 | + | 7 17 2 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 14 19 1 0 0 0 0 | + | 14 19 1 0 0 0 0 |
| − | 4 20 1 0 0 0 0 | + | 4 20 1 0 0 0 0 |
| − | 3 21 1 0 0 0 0 | + | 3 21 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 22 30 1 0 0 0 0 | + | 22 30 1 0 0 0 0 |
| − | 23 31 1 0 0 0 0 | + | 23 31 1 0 0 0 0 |
| − | 24 32 1 0 0 0 0 | + | 24 32 1 0 0 0 0 |
| − | 25 18 1 0 0 0 0 | + | 25 18 1 0 0 0 0 |
| − | 33 34 2 0 0 0 0 | + | 33 34 2 0 0 0 0 |
| − | 33 35 1 0 0 0 0 | + | 33 35 1 0 0 0 0 |
| − | 35 36 2 0 0 0 0 | + | 35 36 2 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 38 2 0 0 0 0 | + | 37 38 2 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 39 40 2 0 0 0 0 | + | 39 40 2 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 41 42 2 0 0 0 0 | + | 41 42 2 0 0 0 0 |
| − | 42 37 1 0 0 0 0 | + | 42 37 1 0 0 0 0 |
| − | 40 43 1 0 0 0 0 | + | 40 43 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 19 44 1 0 0 0 0 | + | 19 44 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL2FEAGS0003 | + | ID FL2FEAGS0003 |
| − | KNApSAcK_ID C00014342 | + | KNApSAcK_ID C00014342 |
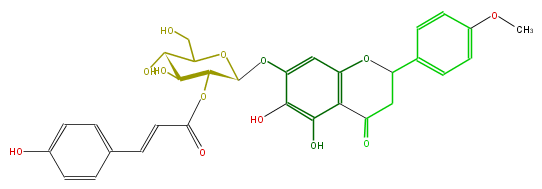
| − | NAME 4'-O-Methylcarthamidin 7-(2-p-coumaroylglucoside);5,6,7-Trihydroxy-4'-methoxyflavanone 7-(2-p-coumaroylglucoside) | + | NAME 4'-O-Methylcarthamidin 7-(2-p-coumaroylglucoside);5,6,7-Trihydroxy-4'-methoxyflavanone 7-(2-p-coumaroylglucoside) |
| − | CAS_RN 292156-63-9 | + | CAS_RN 292156-63-9 |
| − | FORMULA C31H30O13 | + | FORMULA C31H30O13 |
| − | EXACTMASS 610.168641046 | + | EXACTMASS 610.168641046 |
| − | AVERAGEMASS 610.5621 | + | AVERAGEMASS 610.5621 |
| − | SMILES C(C(c(c5)ccc(c5)OC)4)C(c(c(O)1)c(O4)cc(OC(C2OC(=O)C=Cc(c3)ccc(c3)O)OC(C(C(O)2)O)CO)c1O)=O | + | SMILES C(C(c(c5)ccc(c5)OC)4)C(c(c(O)1)c(O4)cc(OC(C2OC(=O)C=Cc(c3)ccc(c3)O)OC(C(C(O)2)O)CO)c1O)=O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
44 48 0 0 0 0 0 0 0 0999 V2000
0.4814 0.6081 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1941 1.0238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4846 -0.2169 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2014 -0.6262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9140 -0.2105 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9104 0.6144 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6303 -0.6199 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3429 -0.2042 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3393 0.6208 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6230 1.0301 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.9931 1.0016 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.7091 0.5919 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.4220 1.0071 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.4188 1.8321 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.7028 2.2419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9899 1.8266 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6314 -1.2631 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1440 0.9692 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
6.0747 2.2108 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2014 -1.3343 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2323 -0.6308 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8158 1.1377 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4032 0.4230 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6048 0.6321 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8113 0.4053 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2239 1.1200 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0223 0.9110 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1793 1.4968 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6591 1.7738 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2990 0.6545 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8518 0.6820 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7763 -0.0078 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1963 -0.8139 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7856 -1.5252 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0202 -0.8139 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4321 -1.5274 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2560 -1.5274 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.6685 -0.8129 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.4935 -0.8129 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.9060 -1.5274 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.4935 -2.2419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.6685 -2.2419 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.7299 -1.5274 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
6.7299 1.8326 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
1 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 2 1 0 0 0 0
5 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 1 0 0 0 0
10 6 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
7 17 2 0 0 0 0
1 18 1 0 0 0 0
14 19 1 0 0 0 0
4 20 1 0 0 0 0
3 21 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
27 28 1 0 0 0 0
28 29 1 0 0 0 0
22 30 1 0 0 0 0
23 31 1 0 0 0 0
24 32 1 0 0 0 0
25 18 1 0 0 0 0
33 34 2 0 0 0 0
33 35 1 0 0 0 0
35 36 2 0 0 0 0
36 37 1 0 0 0 0
37 38 2 0 0 0 0
38 39 1 0 0 0 0
39 40 2 0 0 0 0
40 41 1 0 0 0 0
41 42 2 0 0 0 0
42 37 1 0 0 0 0
40 43 1 0 0 0 0
32 33 1 0 0 0 0
19 44 1 0 0 0 0
S SKP 8
ID FL2FEAGS0003
KNApSAcK_ID C00014342
NAME 4'-O-Methylcarthamidin 7-(2-p-coumaroylglucoside);5,6,7-Trihydroxy-4'-methoxyflavanone 7-(2-p-coumaroylglucoside)
CAS_RN 292156-63-9
FORMULA C31H30O13
EXACTMASS 610.168641046
AVERAGEMASS 610.5621
SMILES C(C(c(c5)ccc(c5)OC)4)C(c(c(O)1)c(O4)cc(OC(C2OC(=O)C=Cc(c3)ccc(c3)O)OC(C(C(O)2)O)CO)c1O)=O
M END