Mol:FL2FAEGS0001
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 44 48 0 0 0 0 0 0 0 0999 V2000 | + | 44 48 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.8011 -0.8644 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8011 -0.8644 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8011 -1.4423 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8011 -1.4423 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3006 -1.7312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3006 -1.7312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1998 -1.4423 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1998 -1.4423 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1998 -0.8644 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1998 -0.8644 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3006 -0.5755 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3006 -0.5755 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7003 -1.7312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7003 -1.7312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2007 -1.4423 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2007 -1.4423 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2007 -0.8644 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2007 -0.8644 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7003 -0.5755 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7003 -0.5755 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7009 -0.5757 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7009 -0.5757 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2152 -0.8726 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2152 -0.8726 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7296 -0.5757 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7296 -0.5757 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7296 0.0182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7296 0.0182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2152 0.3152 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2152 0.3152 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7009 0.0182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7009 0.0182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2007 -0.1838 0.0000 H 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2007 -0.1838 0.0000 H 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3006 -2.3087 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3006 -2.3087 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3012 -0.5757 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3012 -0.5757 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8127 -0.8710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8127 -0.8710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5854 -0.7819 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5854 -0.7819 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1203 -1.2470 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1203 -1.2470 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5489 -0.9213 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5489 -0.9213 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3563 -0.4562 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3563 -0.4562 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9278 -0.7819 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9278 -0.7819 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0953 -1.3749 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0953 -1.3749 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9241 0.1036 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9241 0.1036 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9573 -1.1538 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9573 -1.1538 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7662 -1.4201 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7662 -1.4201 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7003 -2.1028 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7003 -2.1028 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0286 1.8168 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0286 1.8168 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3842 1.2009 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3842 1.2009 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7003 1.3963 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7003 1.3963 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0124 1.2009 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0124 1.2009 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6567 1.8168 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6567 1.8168 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3406 1.6214 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3406 1.6214 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2950 1.2009 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2950 1.2009 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7003 2.1056 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7003 2.1056 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2065 1.6962 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2065 1.6962 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9438 2.3087 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9438 2.3087 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0404 0.6055 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0404 0.6055 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2428 -0.8720 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2428 -0.8720 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2428 0.3145 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2428 0.3145 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9573 -0.0980 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9573 -0.0980 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 9 11 1 0 0 0 0 | + | 9 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 9 17 1 1 0 0 0 | + | 9 17 1 1 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 19 20 1 0 0 0 0 | + | 19 20 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 20 23 1 1 0 0 0 | + | 20 23 1 1 0 0 0 |
| − | 20 24 1 0 0 0 0 | + | 20 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 21 1 0 0 0 0 | + | 25 21 1 0 0 0 0 |
| − | 23 26 1 0 0 0 0 | + | 23 26 1 0 0 0 0 |
| − | 25 27 1 0 0 0 0 | + | 25 27 1 0 0 0 0 |
| − | 21 28 1 0 0 0 0 | + | 21 28 1 0 0 0 0 |
| − | 22 29 1 0 0 0 0 | + | 22 29 1 0 0 0 0 |
| − | 7 30 2 0 0 0 0 | + | 7 30 2 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 35 34 1 1 0 0 0 | + | 35 34 1 1 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 34 37 1 0 0 0 0 | + | 34 37 1 0 0 0 0 |
| − | 33 38 1 0 0 0 0 | + | 33 38 1 0 0 0 0 |
| − | 35 39 1 0 0 0 0 | + | 35 39 1 0 0 0 0 |
| − | 36 40 1 0 0 0 0 | + | 36 40 1 0 0 0 0 |
| − | 32 41 1 0 0 0 0 | + | 32 41 1 0 0 0 0 |
| − | 41 27 1 0 0 0 0 | + | 41 27 1 0 0 0 0 |
| − | 13 42 1 0 0 0 0 | + | 13 42 1 0 0 0 0 |
| − | 14 43 1 0 0 0 0 | + | 14 43 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 43 44 | + | M SAL 1 2 43 44 |
| − | M SBL 1 1 47 | + | M SBL 1 1 47 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 47 -5.4090 3.3919 | + | M SBV 1 47 -5.4090 3.3919 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL2FAEGS0001 | + | ID FL2FAEGS0001 |
| − | KNApSAcK_ID C00000970 | + | KNApSAcK_ID C00000970 |
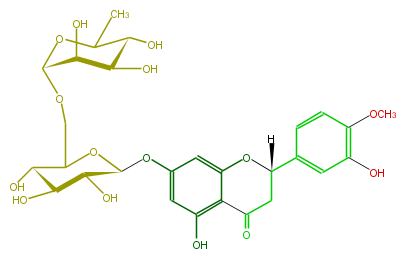
| − | NAME Hesperidin | + | NAME Hesperidin |
| − | CAS_RN 520-26-3 | + | CAS_RN 520-26-3 |
| − | FORMULA C28H34O15 | + | FORMULA C28H34O15 |
| − | EXACTMASS 610.189770418 | + | EXACTMASS 610.189770418 |
| − | AVERAGEMASS 610.56056 | + | AVERAGEMASS 610.56056 |
| − | SMILES c(c1)c(OC)c(O)cc1C([H])(O5)CC(c(c25)c(cc(OC(O3)C(O)C(O)C(O)C3COC(O4)C(C(C(O)C(C)4)O)O)c2)O)=O | + | SMILES c(c1)c(OC)c(O)cc1C([H])(O5)CC(c(c25)c(cc(OC(O3)C(O)C(O)C(O)C3COC(O4)C(C(C(O)C(C)4)O)O)c2)O)=O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
44 48 0 0 0 0 0 0 0 0999 V2000
-0.8011 -0.8644 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8011 -1.4423 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3006 -1.7312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1998 -1.4423 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1998 -0.8644 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3006 -0.5755 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7003 -1.7312 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2007 -1.4423 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2007 -0.8644 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7003 -0.5755 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7009 -0.5757 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2152 -0.8726 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7296 -0.5757 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7296 0.0182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2152 0.3152 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7009 0.0182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2007 -0.1838 0.0000 H 0 0 0 0 0 0 0 0 0 0 0 0
-0.3006 -2.3087 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3012 -0.5757 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8127 -0.8710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5854 -0.7819 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1203 -1.2470 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5489 -0.9213 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3563 -0.4562 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9278 -0.7819 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0953 -1.3749 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9241 0.1036 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9573 -1.1538 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7662 -1.4201 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7003 -2.1028 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0286 1.8168 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3842 1.2009 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7003 1.3963 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0124 1.2009 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6567 1.8168 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3406 1.6214 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2950 1.2009 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7003 2.1056 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2065 1.6962 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9438 2.3087 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0404 0.6055 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2428 -0.8720 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2428 0.3145 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.9573 -0.0980 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
9 17 1 1 0 0 0
3 18 1 0 0 0 0
1 19 1 0 0 0 0
19 20 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
20 23 1 1 0 0 0
20 24 1 0 0 0 0
24 25 1 0 0 0 0
25 21 1 0 0 0 0
23 26 1 0 0 0 0
25 27 1 0 0 0 0
21 28 1 0 0 0 0
22 29 1 0 0 0 0
7 30 2 0 0 0 0
31 32 1 0 0 0 0
32 33 1 1 0 0 0
33 34 1 1 0 0 0
35 34 1 1 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
34 37 1 0 0 0 0
33 38 1 0 0 0 0
35 39 1 0 0 0 0
36 40 1 0 0 0 0
32 41 1 0 0 0 0
41 27 1 0 0 0 0
13 42 1 0 0 0 0
14 43 1 0 0 0 0
43 44 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 43 44
M SBL 1 1 47
M SMT 1 OCH3
M SBV 1 47 -5.4090 3.3919
S SKP 8
ID FL2FAEGS0001
KNApSAcK_ID C00000970
NAME Hesperidin
CAS_RN 520-26-3
FORMULA C28H34O15
EXACTMASS 610.189770418
AVERAGEMASS 610.56056
SMILES c(c1)c(OC)c(O)cc1C([H])(O5)CC(c(c25)c(cc(OC(O3)C(O)C(O)C(O)C3COC(O4)C(C(C(O)C(C)4)O)O)c2)O)=O
M END