Mol:FL2FADGS0001
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 42 46 0 0 0 0 0 0 0 0999 V2000 | + | 42 46 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.5500 -1.5049 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5500 -1.5049 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1645 -1.9174 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1645 -1.9174 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8790 -1.5049 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8790 -1.5049 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8790 -0.6799 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8790 -0.6799 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1645 -0.2675 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1645 -0.2675 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5500 -0.6799 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5500 -0.6799 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5932 -1.9174 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5932 -1.9174 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3077 -1.5049 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3077 -1.5049 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3077 -0.6799 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3077 -0.6799 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5932 -0.2675 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5932 -0.2675 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5932 -2.6361 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5932 -2.6361 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1645 -2.7415 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1645 -2.7415 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2449 -0.2787 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2449 -0.2787 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1029 -0.2693 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1029 -0.2693 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8282 -0.6879 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8282 -0.6879 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5533 -0.2693 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5533 -0.2693 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5533 0.5681 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5533 0.5681 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8282 0.9869 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8282 0.9869 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1029 0.5681 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1029 0.5681 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0930 0.8797 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0930 0.8797 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2569 -0.0891 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2569 -0.0891 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6783 -0.8526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6783 -0.8526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8454 -0.5286 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8454 -0.5286 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9773 -0.5383 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9773 -0.5383 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6258 0.0640 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6258 0.0640 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4766 -0.2414 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4766 -0.2414 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4951 -0.7304 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4951 -0.7304 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1156 -1.0382 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1156 -1.0382 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3799 -1.7298 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3799 -1.7298 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7512 -2.2742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7512 -2.2742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5963 -2.2504 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5963 -2.2504 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1168 -1.7014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1168 -1.7014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1957 -1.3176 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1957 -1.3176 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7558 -2.8534 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7558 -2.8534 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5864 -2.8926 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5864 -2.8926 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9117 -0.2704 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9117 -0.2704 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7452 -1.7636 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7452 -1.7636 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0930 -1.5093 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0930 -1.5093 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8282 1.6245 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8282 1.6245 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3506 2.8926 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3506 2.8926 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2361 0.3247 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2361 0.3247 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3347 1.4521 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3347 1.4521 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 3 7 1 0 0 0 0 | + | 3 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 4 1 0 0 0 0 | + | 10 4 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 2 12 1 0 0 0 0 | + | 2 12 1 0 0 0 0 |
| − | 6 13 1 0 0 0 0 | + | 6 13 1 0 0 0 0 |
| − | 9 14 1 0 0 0 0 | + | 9 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 18 1 0 0 0 0 | + | 17 18 1 0 0 0 0 |
| − | 18 19 2 0 0 0 0 | + | 18 19 2 0 0 0 0 |
| − | 19 14 1 0 0 0 0 | + | 19 14 1 0 0 0 0 |
| − | 17 20 1 0 0 0 0 | + | 17 20 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 22 27 1 0 0 0 0 | + | 22 27 1 0 0 0 0 |
| − | 23 28 1 0 0 0 0 | + | 23 28 1 0 0 0 0 |
| − | 24 13 1 0 0 0 0 | + | 24 13 1 0 0 0 0 |
| − | 29 30 1 1 0 0 0 | + | 29 30 1 1 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 32 31 1 1 0 0 0 | + | 32 31 1 1 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 29 1 0 0 0 0 | + | 33 29 1 0 0 0 0 |
| − | 30 34 1 0 0 0 0 | + | 30 34 1 0 0 0 0 |
| − | 31 35 1 0 0 0 0 | + | 31 35 1 0 0 0 0 |
| − | 32 28 1 0 0 0 0 | + | 32 28 1 0 0 0 0 |
| − | 21 36 1 0 0 0 0 | + | 21 36 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 30 37 1 0 0 0 0 | + | 30 37 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 18 39 1 0 0 0 0 | + | 18 39 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 26 41 1 0 0 0 0 | + | 26 41 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 37 38 | + | M SAL 1 2 37 38 |
| − | M SBL 1 1 42 | + | M SBL 1 1 42 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 42 -0.0060 -0.5106 | + | M SBV 1 42 -0.0060 -0.5106 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 39 40 | + | M SAL 2 2 39 40 |
| − | M SBL 2 1 44 | + | M SBL 2 1 44 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SBV 2 44 0.0000 -0.6376 | + | M SBV 2 44 0.0000 -0.6376 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 41 42 | + | M SAL 3 2 41 42 |
| − | M SBL 3 1 46 | + | M SBL 3 1 46 |
| − | M SMT 3 ^ CH2OH | + | M SMT 3 ^ CH2OH |
| − | M SBV 3 46 0.7595 -0.5661 | + | M SBV 3 46 0.7595 -0.5661 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL2FADGS0001 | + | ID FL2FADGS0001 |
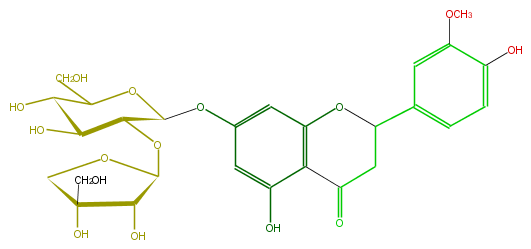
| − | FORMULA C27H32O15 | + | FORMULA C27H32O15 |
| − | EXACTMASS 596.174120354 | + | EXACTMASS 596.174120354 |
| − | AVERAGEMASS 596.5339799999999 | + | AVERAGEMASS 596.5339799999999 |
| − | SMILES c(c1)c(c(cc(C(C5)Oc(c2C(=O)5)cc(OC(O3)C(OC(O4)C(O)C(O)(C4)CO)C(O)C(C(CO)3)O)cc2O)1)OC)O | + | SMILES c(c1)c(c(cc(C(C5)Oc(c2C(=O)5)cc(OC(O3)C(OC(O4)C(O)C(O)(C4)CO)C(O)C(C(CO)3)O)cc2O)1)OC)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
42 46 0 0 0 0 0 0 0 0999 V2000
-0.5500 -1.5049 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1645 -1.9174 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8790 -1.5049 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8790 -0.6799 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1645 -0.2675 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5500 -0.6799 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5932 -1.9174 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3077 -1.5049 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3077 -0.6799 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5932 -0.2675 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5932 -2.6361 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1645 -2.7415 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2449 -0.2787 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1029 -0.2693 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8282 -0.6879 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5533 -0.2693 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5533 0.5681 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8282 0.9869 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1029 0.5681 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.0930 0.8797 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2569 -0.0891 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6783 -0.8526 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8454 -0.5286 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9773 -0.5383 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6258 0.0640 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4766 -0.2414 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4951 -0.7304 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1156 -1.0382 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.3799 -1.7298 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7512 -2.2742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5963 -2.2504 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1168 -1.7014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1957 -1.3176 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7558 -2.8534 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5864 -2.8926 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.9117 -0.2704 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7452 -1.7636 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.0930 -1.5093 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.8282 1.6245 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.3506 2.8926 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2361 0.3247 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3347 1.4521 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
3 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 1 0 0 0 0
10 4 1 0 0 0 0
7 11 2 0 0 0 0
2 12 1 0 0 0 0
6 13 1 0 0 0 0
9 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 18 1 0 0 0 0
18 19 2 0 0 0 0
19 14 1 0 0 0 0
17 20 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
22 27 1 0 0 0 0
23 28 1 0 0 0 0
24 13 1 0 0 0 0
29 30 1 1 0 0 0
30 31 1 1 0 0 0
32 31 1 1 0 0 0
32 33 1 0 0 0 0
33 29 1 0 0 0 0
30 34 1 0 0 0 0
31 35 1 0 0 0 0
32 28 1 0 0 0 0
21 36 1 0 0 0 0
37 38 1 0 0 0 0
30 37 1 0 0 0 0
39 40 1 0 0 0 0
18 39 1 0 0 0 0
41 42 1 0 0 0 0
26 41 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 37 38
M SBL 1 1 42
M SMT 1 CH2OH
M SBV 1 42 -0.0060 -0.5106
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 39 40
M SBL 2 1 44
M SMT 2 OCH3
M SBV 2 44 0.0000 -0.6376
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 41 42
M SBL 3 1 46
M SMT 3 ^ CH2OH
M SBV 3 46 0.7595 -0.5661
S SKP 5
ID FL2FADGS0001
FORMULA C27H32O15
EXACTMASS 596.174120354
AVERAGEMASS 596.5339799999999
SMILES c(c1)c(c(cc(C(C5)Oc(c2C(=O)5)cc(OC(O3)C(OC(O4)C(O)C(O)(C4)CO)C(O)C(C(CO)3)O)cc2O)1)OC)O
M END