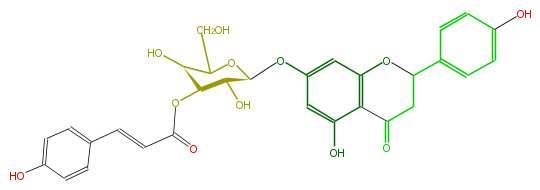
Mol:FL2FAAGS0012
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 42 46 0 0 0 0 0 0 0 0999 V2000 | + | 42 46 0 0 0 0 0 0 0 0999 V2000 |
| − | 0.9737 0.1513 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9737 0.1513 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9737 -0.4911 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9737 -0.4911 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5300 -0.8123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5300 -0.8123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0863 -0.4911 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0863 -0.4911 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0863 0.1513 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0863 0.1513 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5300 0.4725 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5300 0.4725 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6426 -0.8123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6426 -0.8123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1989 -0.4911 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1989 -0.4911 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1989 0.1513 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 3.1989 0.1513 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 2.6426 0.4725 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6426 0.4725 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7550 0.4723 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7550 0.4723 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3220 0.1450 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3220 0.1450 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8890 0.4723 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8890 0.4723 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8890 1.1270 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8890 1.1270 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3220 1.4544 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3220 1.4544 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7550 1.1270 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7550 1.1270 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6426 -1.3560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6426 -1.3560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4174 0.4725 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4174 0.4725 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.4560 1.4544 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.4560 1.4544 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5300 -1.4544 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5300 -1.4544 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6466 0.3391 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -1.6466 0.3391 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -1.2784 -0.1469 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.2784 -0.1469 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -0.7483 0.0593 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -0.7483 0.0593 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -0.2367 0.0648 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -0.2367 0.0648 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -0.6085 0.4366 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6085 0.4366 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0723 0.1918 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -1.0723 0.1918 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -2.1561 0.6332 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1561 0.6332 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7867 -0.4146 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7867 -0.4146 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4445 -0.4506 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4445 -0.4506 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8449 -1.0796 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8449 -1.0796 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4173 -1.3790 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4173 -1.3790 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4493 -1.3614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4493 -1.3614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9955 -0.9790 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9955 -0.9790 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6000 -1.2608 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6000 -1.2608 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0379 -0.9543 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0379 -0.9543 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5224 -1.1802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5224 -1.1802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5690 -1.7127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5690 -1.7127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1311 -2.0192 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1311 -2.0192 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6466 -1.7933 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6466 -1.7933 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0525 -1.9381 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0525 -1.9381 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2736 1.1310 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2736 1.1310 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5316 1.8014 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5316 1.8014 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 9 11 1 0 0 0 0 | + | 9 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 7 17 2 0 0 0 0 | + | 7 17 2 0 0 0 0 |
| − | 18 1 1 0 0 0 0 | + | 18 1 1 0 0 0 0 |
| − | 14 19 1 0 0 0 0 | + | 14 19 1 0 0 0 0 |
| − | 3 20 1 0 0 0 0 | + | 3 20 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 18 1 0 0 0 0 | + | 24 18 1 0 0 0 0 |
| − | 28 30 1 0 0 0 0 | + | 28 30 1 0 0 0 0 |
| − | 30 31 2 0 0 0 0 | + | 30 31 2 0 0 0 0 |
| − | 30 32 1 0 0 0 0 | + | 30 32 1 0 0 0 0 |
| − | 32 33 2 0 0 0 0 | + | 32 33 2 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 2 0 0 0 0 | + | 34 35 2 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 2 0 0 0 0 | + | 36 37 2 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 2 0 0 0 0 | + | 38 39 2 0 0 0 0 |
| − | 39 34 1 0 0 0 0 | + | 39 34 1 0 0 0 0 |
| − | 37 40 1 0 0 0 0 | + | 37 40 1 0 0 0 0 |
| − | 26 41 1 0 0 0 0 | + | 26 41 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 41 42 | + | M SAL 1 2 41 42 |
| − | M SBL 1 1 45 | + | M SBL 1 1 45 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SVB 1 45 -1.2736 1.131 | + | M SVB 1 45 -1.2736 1.131 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL2FAAGS0012 | + | ID FL2FAAGS0012 |
| − | KNApSAcK_ID C00008212 | + | KNApSAcK_ID C00008212 |
| − | NAME Prunin 3''-p-coumarate | + | NAME Prunin 3''-p-coumarate |
| − | CAS_RN 84813-73-0 | + | CAS_RN 84813-73-0 |
| − | FORMULA C30H28O12 | + | FORMULA C30H28O12 |
| − | EXACTMASS 580.15807636 | + | EXACTMASS 580.15807636 |
| − | AVERAGEMASS 580.53612 | + | AVERAGEMASS 580.53612 |
| − | SMILES c(C=CC(=O)O[C@H]([C@@H]2O)[C@H](C(CO)O[C@@H](Oc(c5)cc(c4c(O)5)OC(CC(=O)4)c(c3)ccc(O)c3)2)O)(c1)ccc(O)c1 | + | SMILES c(C=CC(=O)O[C@H]([C@@H]2O)[C@H](C(CO)O[C@@H](Oc(c5)cc(c4c(O)5)OC(CC(=O)4)c(c3)ccc(O)c3)2)O)(c1)ccc(O)c1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
42 46 0 0 0 0 0 0 0 0999 V2000
0.9737 0.1513 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9737 -0.4911 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5300 -0.8123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0863 -0.4911 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0863 0.1513 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5300 0.4725 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6426 -0.8123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1989 -0.4911 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1989 0.1513 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
2.6426 0.4725 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7550 0.4723 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3220 0.1450 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.8890 0.4723 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.8890 1.1270 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3220 1.4544 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7550 1.1270 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6426 -1.3560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4174 0.4725 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.4560 1.4544 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5300 -1.4544 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6466 0.3391 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-1.2784 -0.1469 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.7483 0.0593 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.2367 0.0648 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.6085 0.4366 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0723 0.1918 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-2.1561 0.6332 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7867 -0.4146 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.4445 -0.4506 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8449 -1.0796 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4173 -1.3790 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4493 -1.3614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9955 -0.9790 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6000 -1.2608 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0379 -0.9543 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.5224 -1.1802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.5690 -1.7127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1311 -2.0192 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6466 -1.7933 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.0525 -1.9381 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2736 1.1310 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5316 1.8014 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
7 17 2 0 0 0 0
18 1 1 0 0 0 0
14 19 1 0 0 0 0
3 20 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
24 18 1 0 0 0 0
28 30 1 0 0 0 0
30 31 2 0 0 0 0
30 32 1 0 0 0 0
32 33 2 0 0 0 0
33 34 1 0 0 0 0
34 35 2 0 0 0 0
35 36 1 0 0 0 0
36 37 2 0 0 0 0
37 38 1 0 0 0 0
38 39 2 0 0 0 0
39 34 1 0 0 0 0
37 40 1 0 0 0 0
26 41 1 0 0 0 0
41 42 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 41 42
M SBL 1 1 45
M SMT 1 CH2OH
M SVB 1 45 -1.2736 1.131
S SKP 8
ID FL2FAAGS0012
KNApSAcK_ID C00008212
NAME Prunin 3''-p-coumarate
CAS_RN 84813-73-0
FORMULA C30H28O12
EXACTMASS 580.15807636
AVERAGEMASS 580.53612
SMILES c(C=CC(=O)O[C@H]([C@@H]2O)[C@H](C(CO)O[C@@H](Oc(c5)cc(c4c(O)5)OC(CC(=O)4)c(c3)ccc(O)c3)2)O)(c1)ccc(O)c1
M END