Mol:FL2FAACS0003
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 42 46 0 0 0 0 0 0 0 0999 V2000 | + | 42 46 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.0054 -0.9431 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0054 -0.9431 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0054 -1.5855 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0054 -1.5855 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4491 -1.9066 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4491 -1.9066 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1072 -1.5855 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1072 -1.5855 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1072 -0.9431 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1072 -0.9431 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4491 -0.6219 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4491 -0.6219 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6635 -1.9066 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6635 -1.9066 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2198 -1.5855 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2198 -1.5855 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2198 -0.9431 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2198 -0.9431 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6635 -0.6219 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6635 -0.6219 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6635 -2.4075 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6635 -2.4075 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5615 -0.6220 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5615 -0.6220 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2037 2.0625 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2037 2.0625 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8093 1.6038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8093 1.6038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5524 0.9431 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5524 0.9431 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5455 0.3057 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5455 0.3057 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0822 0.7689 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0822 0.7689 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3872 1.3469 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3872 1.3469 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6023 2.7529 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6023 2.7529 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8441 2.3190 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8441 2.3190 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1878 0.5646 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1878 0.5646 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4491 -2.5488 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4491 -2.5488 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8378 -0.6014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8378 -0.6014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4240 -0.9399 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4240 -0.9399 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0102 -0.6014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0102 -0.6014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0102 0.0755 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0102 0.0755 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4240 0.4139 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4240 0.4139 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8378 0.0755 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8378 0.0755 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5961 0.4137 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5961 0.4137 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0822 -1.9567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0822 -1.9567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7111 -2.4467 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7111 -2.4467 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1766 -2.2388 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1766 -2.2388 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6608 -2.2332 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6608 -2.2332 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0356 -1.8584 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0356 -1.8584 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5032 -2.1052 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5032 -2.1052 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5961 -2.2534 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5961 -2.2534 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2897 -2.4748 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2897 -2.4748 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8703 -2.7529 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8703 -2.7529 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0364 1.5914 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0364 1.5914 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7509 1.1789 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7509 1.1789 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7598 -1.1071 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7598 -1.1071 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0453 -1.5196 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0453 -1.5196 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 1 12 1 0 0 0 0 | + | 1 12 1 0 0 0 0 |
| − | 13 14 1 1 0 0 0 | + | 13 14 1 1 0 0 0 |
| − | 14 15 1 1 0 0 0 | + | 14 15 1 1 0 0 0 |
| − | 16 15 1 1 0 0 0 | + | 16 15 1 1 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 18 1 0 0 0 0 | + | 17 18 1 0 0 0 0 |
| − | 18 13 1 0 0 0 0 | + | 18 13 1 0 0 0 0 |
| − | 13 19 1 0 0 0 0 | + | 13 19 1 0 0 0 0 |
| − | 14 20 1 0 0 0 0 | + | 14 20 1 0 0 0 0 |
| − | 15 21 1 0 0 0 0 | + | 15 21 1 0 0 0 0 |
| − | 6 16 1 0 0 0 0 | + | 6 16 1 0 0 0 0 |
| − | 3 22 1 0 0 0 0 | + | 3 22 1 0 0 0 0 |
| − | 23 24 2 0 0 0 0 | + | 23 24 2 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 2 0 0 0 0 | + | 25 26 2 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 2 0 0 0 0 | + | 27 28 2 0 0 0 0 |
| − | 28 23 1 0 0 0 0 | + | 28 23 1 0 0 0 0 |
| − | 23 9 1 0 0 0 0 | + | 23 9 1 0 0 0 0 |
| − | 26 29 1 0 0 0 0 | + | 26 29 1 0 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 33 32 1 1 0 0 0 | + | 33 32 1 1 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 30 1 0 0 0 0 | + | 35 30 1 0 0 0 0 |
| − | 30 36 1 0 0 0 0 | + | 30 36 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 2 1 0 0 0 0 | + | 33 2 1 0 0 0 0 |
| − | 18 39 1 0 0 0 0 | + | 18 39 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 35 41 1 0 0 0 0 | + | 35 41 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 39 40 | + | M SAL 1 2 39 40 |
| − | M SBL 1 1 43 | + | M SBL 1 1 43 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 43 -6.2289 5.6127 | + | M SBV 1 43 -6.2289 5.6127 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 41 42 | + | M SAL 2 2 41 42 |
| − | M SBL 2 1 45 | + | M SBL 2 1 45 |
| − | M SMT 2 ^CH2OH | + | M SMT 2 ^CH2OH |
| − | M SBV 2 45 -6.9091 6.3662 | + | M SBV 2 45 -6.9091 6.3662 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL2FAACS0003 | + | ID FL2FAACS0003 |
| − | KNApSAcK_ID C00006398 | + | KNApSAcK_ID C00006398 |
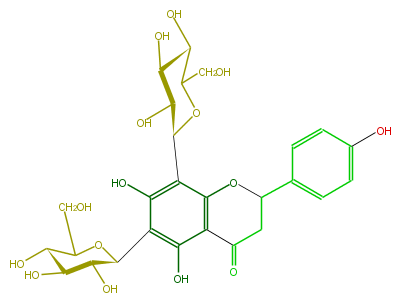
| − | NAME 6,8-Di-C-glucopyranosylnaringenin | + | NAME 6,8-Di-C-glucopyranosylnaringenin |
| − | CAS_RN 81426-09-7 | + | CAS_RN 81426-09-7 |
| − | FORMULA C27H32O15 | + | FORMULA C27H32O15 |
| − | EXACTMASS 596.174120354 | + | EXACTMASS 596.174120354 |
| − | AVERAGEMASS 596.5339799999999 | + | AVERAGEMASS 596.5339799999999 |
| − | SMILES C(C1O)(OC(c(c35)c(c(c(O)c3C(=O)CC(O5)c(c4)ccc(O)c4)C(O2)C(C(C(C(CO)2)O)O)O)O)C(C1O)O)CO | + | SMILES C(C1O)(OC(c(c35)c(c(c(O)c3C(=O)CC(O5)c(c4)ccc(O)c4)C(O2)C(C(C(C(CO)2)O)O)O)O)C(C1O)O)CO |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
42 46 0 0 0 0 0 0 0 0999 V2000
-1.0054 -0.9431 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0054 -1.5855 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4491 -1.9066 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1072 -1.5855 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1072 -0.9431 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4491 -0.6219 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6635 -1.9066 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2198 -1.5855 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2198 -0.9431 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6635 -0.6219 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6635 -2.4075 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.5615 -0.6220 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2037 2.0625 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8093 1.6038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5524 0.9431 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5455 0.3057 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0822 0.7689 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.3872 1.3469 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6023 2.7529 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8441 2.3190 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.1878 0.5646 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.4491 -2.5488 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8378 -0.6014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4240 -0.9399 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0102 -0.6014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0102 0.0755 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4240 0.4139 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8378 0.0755 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5961 0.4137 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0822 -1.9567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7111 -2.4467 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1766 -2.2388 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6608 -2.2332 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0356 -1.8584 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5032 -2.1052 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5961 -2.2534 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2897 -2.4748 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8703 -2.7529 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0364 1.5914 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7509 1.1789 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7598 -1.1071 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0453 -1.5196 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
1 12 1 0 0 0 0
13 14 1 1 0 0 0
14 15 1 1 0 0 0
16 15 1 1 0 0 0
16 17 1 0 0 0 0
17 18 1 0 0 0 0
18 13 1 0 0 0 0
13 19 1 0 0 0 0
14 20 1 0 0 0 0
15 21 1 0 0 0 0
6 16 1 0 0 0 0
3 22 1 0 0 0 0
23 24 2 0 0 0 0
24 25 1 0 0 0 0
25 26 2 0 0 0 0
26 27 1 0 0 0 0
27 28 2 0 0 0 0
28 23 1 0 0 0 0
23 9 1 0 0 0 0
26 29 1 0 0 0 0
30 31 1 1 0 0 0
31 32 1 1 0 0 0
33 32 1 1 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 30 1 0 0 0 0
30 36 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 2 1 0 0 0 0
18 39 1 0 0 0 0
39 40 1 0 0 0 0
35 41 1 0 0 0 0
41 42 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 39 40
M SBL 1 1 43
M SMT 1 CH2OH
M SBV 1 43 -6.2289 5.6127
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 41 42
M SBL 2 1 45
M SMT 2 ^CH2OH
M SBV 2 45 -6.9091 6.3662
S SKP 8
ID FL2FAACS0003
KNApSAcK_ID C00006398
NAME 6,8-Di-C-glucopyranosylnaringenin
CAS_RN 81426-09-7
FORMULA C27H32O15
EXACTMASS 596.174120354
AVERAGEMASS 596.5339799999999
SMILES C(C1O)(OC(c(c35)c(c(c(O)c3C(=O)CC(O5)c(c4)ccc(O)c4)C(O2)C(C(C(C(CO)2)O)O)O)O)C(C1O)O)CO
M END