Mol:FL1CDAGS0002
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 42 45 0 0 0 0 0 0 0 0999 V2000 | + | 42 45 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.7714 -0.7554 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7714 -0.7554 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7714 -1.5807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7714 -1.5807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0566 -1.9932 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0566 -1.9932 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3419 -1.5807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3419 -1.5807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3419 -0.7554 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3419 -0.7554 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0566 -0.3426 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0566 -0.3426 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6274 -1.9931 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6274 -1.9931 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9146 -1.5816 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9146 -1.5816 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2032 -1.9922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2032 -1.9922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5066 -1.5825 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5066 -1.5825 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2012 -1.9835 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2012 -1.9835 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8959 -1.5825 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8959 -1.5825 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8959 -0.7804 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8959 -0.7804 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2012 -0.3794 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2012 -0.3794 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5066 -0.7804 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5066 -0.7804 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6274 -2.8163 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6274 -2.8163 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0566 -2.8183 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0566 -2.8183 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5592 -0.3878 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5592 -0.3878 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8009 0.6088 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8009 0.6088 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6466 0.6088 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6466 0.6088 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0388 1.1967 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0388 1.1967 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8310 2.0214 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8310 2.0214 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9853 2.0214 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9853 2.0214 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5931 1.4333 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5931 1.4333 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7718 2.8183 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7718 2.8183 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3934 2.6033 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3934 2.6033 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8403 1.1632 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8403 1.1632 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6351 1.2438 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6351 1.2438 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6351 1.7611 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6351 1.7611 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4187 1.7836 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4187 1.7836 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1027 1.8820 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1027 1.8820 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8426 1.8820 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8426 1.8820 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4187 1.2438 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4187 1.2438 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9734 2.1868 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9734 2.1868 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7036 1.8076 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7036 1.8076 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3074 0.5803 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3074 0.5803 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4187 0.7685 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4187 0.7685 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7043 0.3560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7043 0.3560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4578 -0.3089 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4578 -0.3089 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.1027 0.8086 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.1027 0.8086 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6041 -0.2424 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6041 -0.2424 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6822 -0.7747 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6822 -0.7747 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 11 2 0 0 0 0 | + | 10 11 2 0 0 0 0 |
| − | 11 12 1 0 0 0 0 | + | 11 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 10 1 0 0 0 0 | + | 15 10 1 0 0 0 0 |
| − | 7 16 2 0 0 0 0 | + | 7 16 2 0 0 0 0 |
| − | 3 17 1 0 0 0 0 | + | 3 17 1 0 0 0 0 |
| − | 20 19 1 1 0 0 0 | + | 20 19 1 1 0 0 0 |
| − | 20 21 1 0 0 0 0 | + | 20 21 1 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 19 1 1 0 0 0 | + | 24 19 1 1 0 0 0 |
| − | 23 25 1 0 0 0 0 | + | 23 25 1 0 0 0 0 |
| − | 22 26 1 0 0 0 0 | + | 22 26 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 20 18 1 0 0 0 0 | + | 20 18 1 0 0 0 0 |
| − | 33 28 1 1 0 0 0 | + | 33 28 1 1 0 0 0 |
| − | 32 28 1 1 0 0 0 | + | 32 28 1 1 0 0 0 |
| − | 31 33 1 1 0 0 0 | + | 31 33 1 1 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 33 30 1 0 0 0 0 | + | 33 30 1 0 0 0 0 |
| − | 34 31 1 0 0 0 0 | + | 34 31 1 0 0 0 0 |
| − | 32 34 1 0 0 0 0 | + | 32 34 1 0 0 0 0 |
| − | 27 32 1 0 0 0 0 | + | 27 32 1 0 0 0 0 |
| − | 18 13 1 0 0 0 0 | + | 18 13 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 24 35 1 0 0 0 0 | + | 24 35 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 33 37 1 0 0 0 0 | + | 33 37 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 1 39 1 0 0 0 0 | + | 1 39 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 5 41 1 0 0 0 0 | + | 5 41 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 35 36 | + | M SAL 1 2 35 36 |
| − | M SBL 1 1 39 | + | M SBL 1 1 39 |
| − | M SMT 1 ^ CH2OH | + | M SMT 1 ^ CH2OH |
| − | M SBV 1 39 0.8896 -0.3743 | + | M SBV 1 39 0.8896 -0.3743 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 37 38 | + | M SAL 2 2 37 38 |
| − | M SBL 2 1 41 | + | M SBL 2 1 41 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SBV 2 41 0.0000 0.4754 | + | M SBV 2 41 0.0000 0.4754 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 39 40 | + | M SAL 3 2 39 40 |
| − | M SBL 3 1 43 | + | M SBL 3 1 43 |
| − | M SMT 3 ^ OCH3 | + | M SMT 3 ^ OCH3 |
| − | M SBV 3 43 0.6864 -0.4464 | + | M SBV 3 43 0.6864 -0.4464 |
| − | M STY 1 4 SUP | + | M STY 1 4 SUP |
| − | M SLB 1 4 4 | + | M SLB 1 4 4 |
| − | M SAL 4 2 41 42 | + | M SAL 4 2 41 42 |
| − | M SBL 4 1 45 | + | M SBL 4 1 45 |
| − | M SMT 4 OCH3 | + | M SMT 4 OCH3 |
| − | M SBV 4 45 -0.7378 -0.5129 | + | M SBV 4 45 -0.7378 -0.5129 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL1CDAGS0002 | + | ID FL1CDAGS0002 |
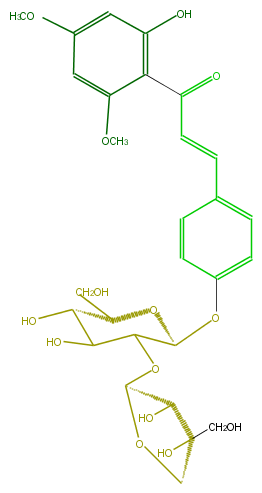
| − | FORMULA C28H34O14 | + | FORMULA C28H34O14 |
| − | EXACTMASS 594.194855796 | + | EXACTMASS 594.194855796 |
| − | AVERAGEMASS 594.56116 | + | AVERAGEMASS 594.56116 |
| − | SMILES C(C(OC(C4O)OCC(CO)4O)3)(OC(CO)C(C(O)3)O)Oc(c2)ccc(c2)C=CC(=O)c(c1O)c(OC)cc(OC)c1 | + | SMILES C(C(OC(C4O)OCC(CO)4O)3)(OC(CO)C(C(O)3)O)Oc(c2)ccc(c2)C=CC(=O)c(c1O)c(OC)cc(OC)c1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
42 45 0 0 0 0 0 0 0 0999 V2000
-3.7714 -0.7554 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7714 -1.5807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0566 -1.9932 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3419 -1.5807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3419 -0.7554 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0566 -0.3426 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6274 -1.9931 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9146 -1.5816 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2032 -1.9922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5066 -1.5825 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2012 -1.9835 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8959 -1.5825 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8959 -0.7804 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2012 -0.3794 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5066 -0.7804 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6274 -2.8163 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0566 -2.8183 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5592 -0.3878 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8009 0.6088 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6466 0.6088 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0388 1.1967 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8310 2.0214 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9853 2.0214 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5931 1.4333 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7718 2.8183 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3934 2.6033 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8403 1.1632 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6351 1.2438 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6351 1.7611 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.4187 1.7836 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.1027 1.8820 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8426 1.8820 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4187 1.2438 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9734 2.1868 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7036 1.8076 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3074 0.5803 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.4187 0.7685 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7043 0.3560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.4578 -0.3089 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.1027 0.8086 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6041 -0.2424 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6822 -0.7747 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 11 2 0 0 0 0
11 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 10 1 0 0 0 0
7 16 2 0 0 0 0
3 17 1 0 0 0 0
20 19 1 1 0 0 0
20 21 1 0 0 0 0
21 22 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 19 1 1 0 0 0
23 25 1 0 0 0 0
22 26 1 0 0 0 0
21 27 1 0 0 0 0
20 18 1 0 0 0 0
33 28 1 1 0 0 0
32 28 1 1 0 0 0
31 33 1 1 0 0 0
28 29 1 0 0 0 0
33 30 1 0 0 0 0
34 31 1 0 0 0 0
32 34 1 0 0 0 0
27 32 1 0 0 0 0
18 13 1 0 0 0 0
35 36 1 0 0 0 0
24 35 1 0 0 0 0
37 38 1 0 0 0 0
33 37 1 0 0 0 0
39 40 1 0 0 0 0
1 39 1 0 0 0 0
41 42 1 0 0 0 0
5 41 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 35 36
M SBL 1 1 39
M SMT 1 ^ CH2OH
M SBV 1 39 0.8896 -0.3743
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 37 38
M SBL 2 1 41
M SMT 2 CH2OH
M SBV 2 41 0.0000 0.4754
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 39 40
M SBL 3 1 43
M SMT 3 ^ OCH3
M SBV 3 43 0.6864 -0.4464
M STY 1 4 SUP
M SLB 1 4 4
M SAL 4 2 41 42
M SBL 4 1 45
M SMT 4 OCH3
M SBV 4 45 -0.7378 -0.5129
S SKP 5
ID FL1CDAGS0002
FORMULA C28H34O14
EXACTMASS 594.194855796
AVERAGEMASS 594.56116
SMILES C(C(OC(C4O)OCC(CO)4O)3)(OC(CO)C(C(O)3)O)Oc(c2)ccc(c2)C=CC(=O)c(c1O)c(OC)cc(OC)c1
M END